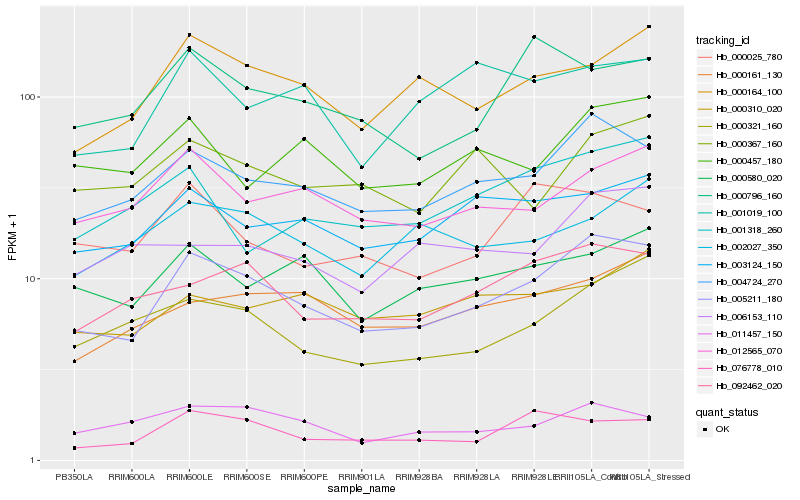
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005211_180 |
0.0 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 2 |
Hb_004724_270 |
0.0801578163 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 3 |
Hb_092462_020 |
0.1124953679 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000321_160 |
0.1191939776 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000164_100 |
0.1208699323 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 6 |
Hb_012565_070 |
0.1217090422 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_003124_150 |
0.1225949664 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 8 |
Hb_000161_130 |
0.1266075678 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_076778_010 |
0.1266981394 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 10 |
Hb_000580_020 |
0.1290461009 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_011457_150 |
0.1320383963 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 12 |
Hb_000457_180 |
0.1326944187 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000025_780 |
0.1347116851 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 14 |
Hb_006153_110 |
0.1374342962 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_000367_160 |
0.1382627781 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |
| 16 |
Hb_001318_260 |
0.1384208772 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 17 |
Hb_000796_160 |
0.1384717064 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000310_020 |
0.1392225068 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 19 |
Hb_002027_350 |
0.139697076 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 20 |
Hb_001019_100 |
0.1404114787 |
- |
- |
BnaC02g13240D [Brassica napus] |