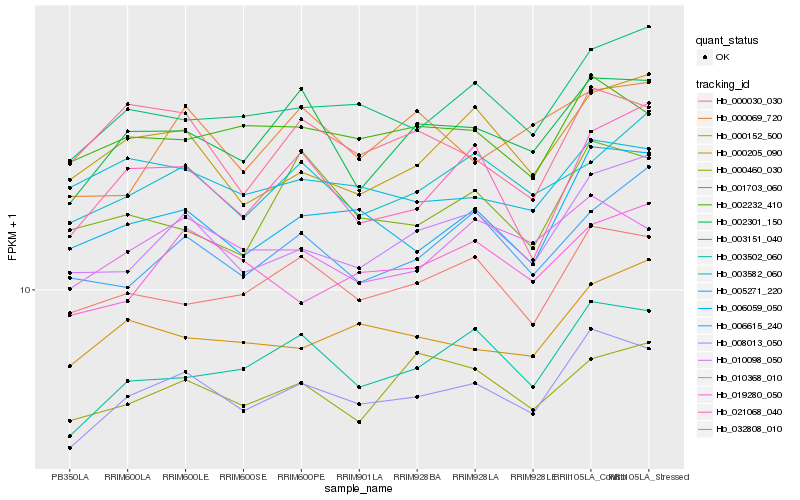
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008013_050 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 2 |
Hb_006059_050 |
0.0592962162 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 3 |
Hb_002301_150 |
0.0641347205 |
- |
- |
Drought-responsive family protein [Theobroma cacao] |
| 4 |
Hb_003502_060 |
0.0641945417 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 5 |
Hb_003151_040 |
0.0662969408 |
transcription factor |
TF Family: Alfin-like |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_010098_050 |
0.0684523364 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 7 |
Hb_000030_030 |
0.0686034006 |
- |
- |
PREDICTED: ADP-glucose phosphorylase [Jatropha curcas] |
| 8 |
Hb_010368_010 |
0.0718823662 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 9 |
Hb_002232_410 |
0.073699565 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 10 |
Hb_000152_500 |
0.0798545405 |
- |
- |
PREDICTED: probable mitochondrial intermediate peptidase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003582_060 |
0.0806619427 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_019280_050 |
0.0806996154 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 2 [Jatropha curcas] |
| 13 |
Hb_000205_090 |
0.0809331325 |
- |
- |
hypothetical protein POPTR_0019s03570g [Populus trichocarpa] |
| 14 |
Hb_000460_030 |
0.0815563179 |
- |
- |
4-hydroxybenzoate octaprenyltransferase, putative [Ricinus communis] |
| 15 |
Hb_021068_040 |
0.081966069 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 16 |
Hb_006615_240 |
0.0852798998 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 17 |
Hb_032808_010 |
0.0854920962 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 18 |
Hb_005271_220 |
0.0858797794 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 19 |
Hb_001703_060 |
0.0872178768 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000069_720 |
0.0879664293 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |