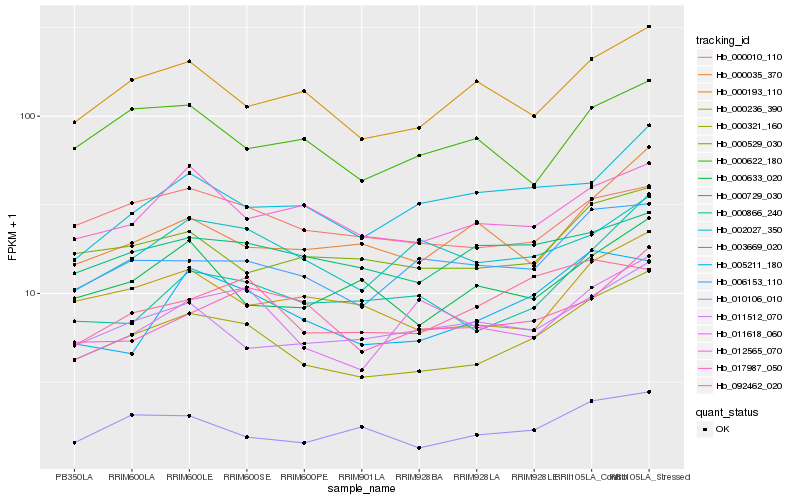
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000321_160 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006153_110 |
0.1074720926 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_000236_390 |
0.1077494237 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 4 |
Hb_000193_110 |
0.1100052651 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 5 |
Hb_000529_030 |
0.1103267112 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_010106_010 |
0.1125913299 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 7 |
Hb_011512_070 |
0.114584931 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 8 |
Hb_005211_180 |
0.1191939776 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 9 |
Hb_002027_350 |
0.1207544928 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 10 |
Hb_000866_240 |
0.1209509345 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 11 |
Hb_000633_020 |
0.1220765229 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 12 |
Hb_012565_070 |
0.1227862316 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 13 |
Hb_000035_370 |
0.1235742166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011618_060 |
0.1241464193 |
- |
- |
PREDICTED: DNA-repair protein XRCC1 [Jatropha curcas] |
| 15 |
Hb_003669_020 |
0.1247340463 |
- |
- |
PREDICTED: high mobility group nucleosome-binding domain-containing protein 5 [Populus euphratica] |
| 16 |
Hb_017987_050 |
0.1257128302 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_092462_020 |
0.1277071364 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000010_110 |
0.1299095492 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000729_030 |
0.1302862471 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 20 |
Hb_000622_180 |
0.1314674183 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |