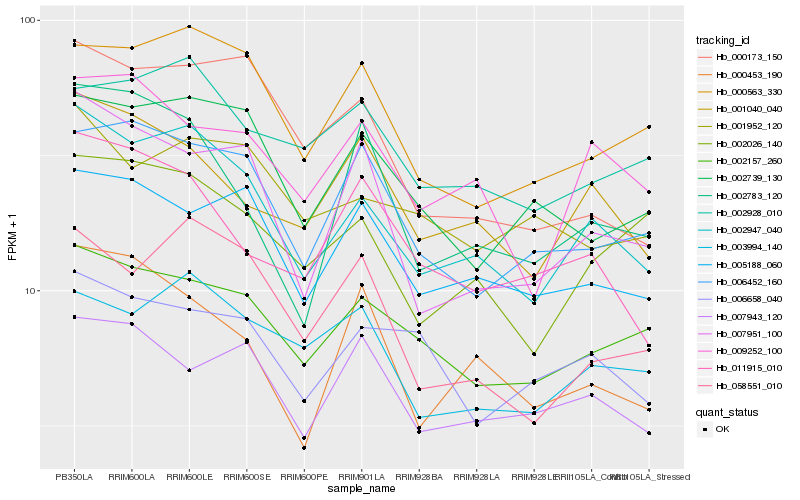
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002947_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |
| 2 |
Hb_002157_260 |
0.1041111887 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 3 |
Hb_001040_040 |
0.1041955115 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 4 |
Hb_007951_100 |
0.1058851065 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_000563_330 |
0.1081201097 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_005188_060 |
0.1084931189 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_009252_100 |
0.1092662669 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 8 |
Hb_058551_010 |
0.1098484968 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_002026_140 |
0.1105342839 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 10 |
Hb_006452_160 |
0.1151520828 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 11 |
Hb_002739_130 |
0.1157524045 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 12 |
Hb_003994_140 |
0.1159175895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000173_150 |
0.1166361862 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 14 |
Hb_011915_010 |
0.1178947984 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 15 |
Hb_002783_120 |
0.1204465286 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 16 |
Hb_000453_190 |
0.1215998253 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 17 |
Hb_001952_120 |
0.1217074999 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002928_010 |
0.1219334524 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 19 |
Hb_006658_040 |
0.1228308852 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 20 |
Hb_007943_120 |
0.1245998389 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |