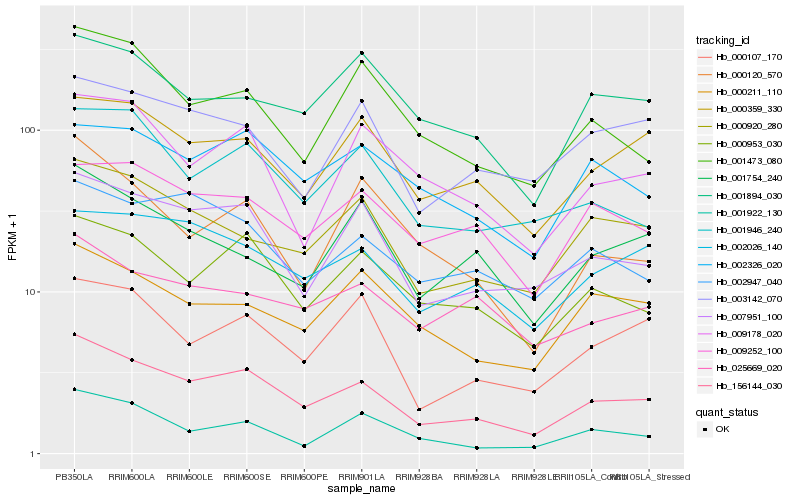
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156144_030 |
0.0 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 2 |
Hb_007951_100 |
0.0957391798 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000953_030 |
0.1019707044 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001946_240 |
0.1146244418 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 5 |
Hb_000920_280 |
0.115495815 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 6 |
Hb_000359_330 |
0.1210721678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001473_080 |
0.121678004 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 8 |
Hb_000107_170 |
0.1228030493 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 9 |
Hb_002026_140 |
0.1240940583 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 10 |
Hb_002947_040 |
0.1246661693 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |
| 11 |
Hb_009178_020 |
0.1250079744 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 12 |
Hb_000211_110 |
0.1295569764 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_009252_100 |
0.1299787898 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 14 |
Hb_002326_020 |
0.13191392 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 15 |
Hb_000120_570 |
0.1325308769 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 16 |
Hb_001894_030 |
0.1333244344 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 17 |
Hb_001754_240 |
0.133754702 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 18 |
Hb_001922_130 |
0.1338240378 |
- |
- |
hypothetical protein PHAVU_002G294300g [Phaseolus vulgaris] |
| 19 |
Hb_003142_070 |
0.1340960492 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 20 |
Hb_025669_020 |
0.1341649121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |