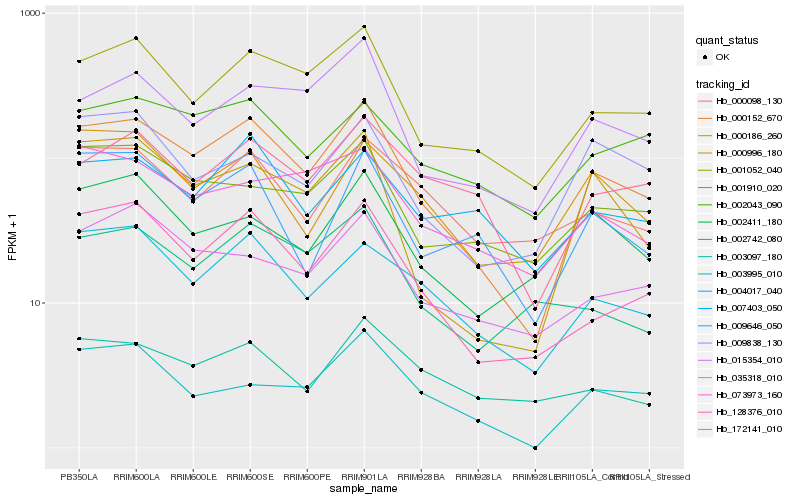
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_670 |
0.0 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 2 |
Hb_001052_040 |
0.1074435343 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_128376_010 |
0.1079018443 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000996_180 |
0.1221392875 |
- |
- |
PREDICTED: tetraspanin-10 [Jatropha curcas] |
| 5 |
Hb_004017_040 |
0.1257650296 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 6 |
Hb_015354_010 |
0.1304341663 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 7 |
Hb_009646_050 |
0.1368023049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000186_260 |
0.1432097391 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 9 |
Hb_035318_010 |
0.1440821587 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 10 |
Hb_001910_020 |
0.1441943032 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_003995_010 |
0.1443735662 |
- |
- |
- |
| 12 |
Hb_172141_010 |
0.1480142535 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |
| 13 |
Hb_002043_090 |
0.1483235731 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 14 |
Hb_002411_180 |
0.1491604547 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000098_130 |
0.1492297769 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 16 |
Hb_002742_080 |
0.1501095857 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_073973_160 |
0.1510303744 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 18 |
Hb_009838_130 |
0.1512963038 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 19 |
Hb_003097_180 |
0.1514160819 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 20 |
Hb_007403_050 |
0.1514809866 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |