| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
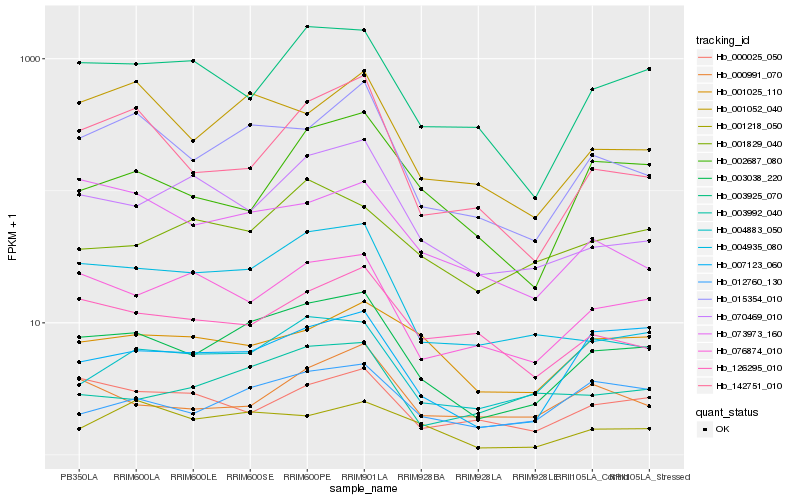
Hb_003038_220 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 2 |
Hb_015354_010 |
0.1192183916 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_012760_130 |
0.1445520965 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 4 |
Hb_007123_060 |
0.1469253856 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 5 |
Hb_003925_070 |
0.1480597069 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 6 |
Hb_004883_050 |
0.1494699959 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 7 |
Hb_004935_080 |
0.14988367 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 8 |
Hb_001052_040 |
0.1501133294 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_076874_010 |
0.1509019277 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 10 |
Hb_003992_040 |
0.1543109851 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 11 |
Hb_070469_010 |
0.1603937609 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 12 |
Hb_142751_010 |
0.1609443959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001218_050 |
0.1640138352 |
- |
- |
Ribosome-binding protein, putative [Ricinus communis] |
| 14 |
Hb_126295_010 |
0.1658397874 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 15 |
Hb_002687_080 |
0.1663060361 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 16 |
Hb_001025_110 |
0.1671105477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000991_070 |
0.1694325552 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 18 |
Hb_001829_040 |
0.1734616991 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 19 |
Hb_073973_160 |
0.1743274704 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 20 |
Hb_000025_050 |
0.1745515957 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |