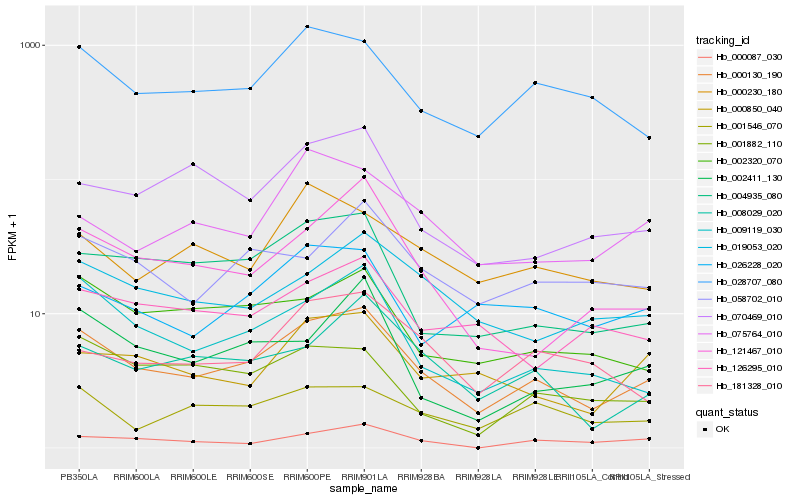
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 2 |
Hb_009119_030 |
0.1315762341 |
- |
- |
GMFP4 [Medicago truncatula] |
| 3 |
Hb_028707_080 |
0.1402393434 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 4 |
Hb_004935_080 |
0.1443987364 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 5 |
Hb_002320_070 |
0.1485331086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008029_020 |
0.1507474183 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 7 |
Hb_019053_020 |
0.1527334378 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 8 |
Hb_121467_010 |
0.1545233911 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 9 |
Hb_058702_010 |
0.1572275279 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_070469_010 |
0.1574016356 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 11 |
Hb_000850_040 |
0.159586231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001546_070 |
0.1610293272 |
- |
- |
Uncharacterized protein F383_19872 [Gossypium arboreum] |
| 13 |
Hb_002411_130 |
0.165658168 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 14 |
Hb_126295_010 |
0.1662187326 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 15 |
Hb_181328_010 |
0.1667765819 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_026228_020 |
0.1670023346 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 17 |
Hb_075764_010 |
0.167251872 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 18 |
Hb_001882_110 |
0.1693339874 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000230_180 |
0.17095232 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_000087_030 |
0.1714641025 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |