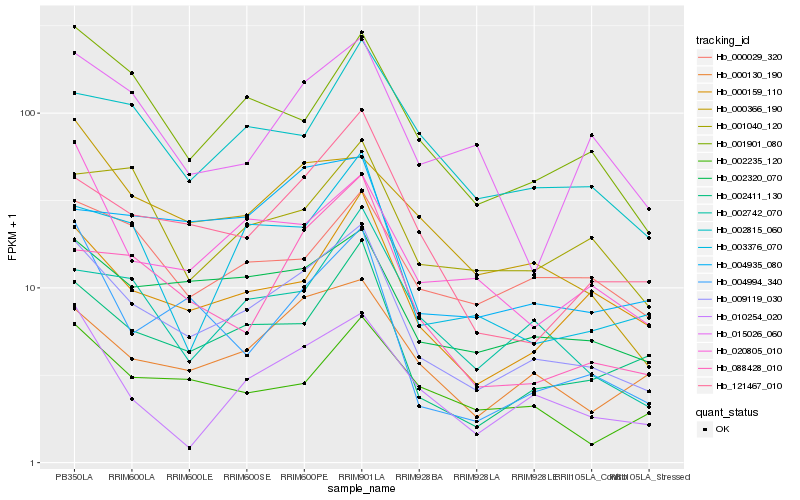
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009119_030 |
0.0 |
- |
- |
GMFP4 [Medicago truncatula] |
| 2 |
Hb_002320_070 |
0.1206095004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002411_130 |
0.126478944 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 4 |
Hb_000130_190 |
0.1315762341 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 5 |
Hb_000159_110 |
0.1342454033 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 6 |
Hb_001901_080 |
0.1357265912 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 7 |
Hb_121467_010 |
0.1418561779 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 8 |
Hb_000366_190 |
0.144189061 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 9 |
Hb_020805_010 |
0.1487197076 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_002742_070 |
0.1494220793 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 11 |
Hb_003376_070 |
0.1501538139 |
- |
- |
hypothetical protein B456_009G419800 [Gossypium raimondii] |
| 12 |
Hb_002815_060 |
0.153143532 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 13 |
Hb_010254_020 |
0.1551648381 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 14 |
Hb_002235_120 |
0.1571336399 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_015026_060 |
0.1585207615 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 16 |
Hb_004935_080 |
0.1596129449 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 17 |
Hb_000029_320 |
0.1602982258 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 18 |
Hb_088428_010 |
0.1603826843 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 19 |
Hb_004994_340 |
0.1604388123 |
- |
- |
- |
| 20 |
Hb_001040_120 |
0.162398915 |
- |
- |
allantoinase, putative [Ricinus communis] |