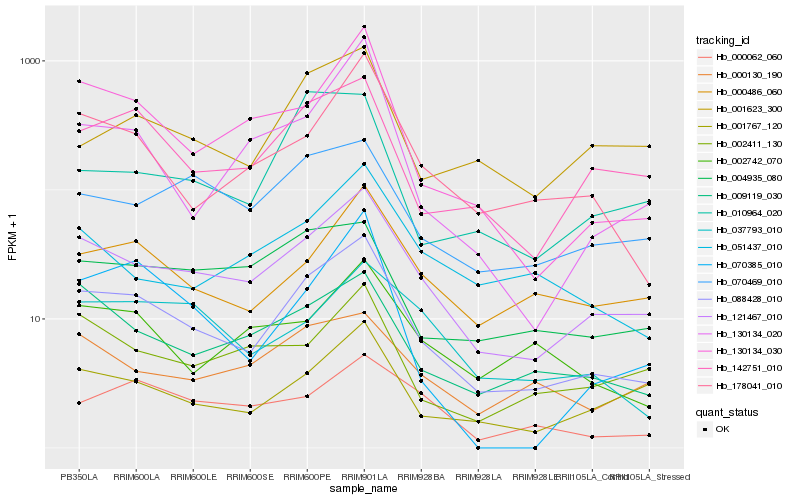
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088428_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 2 |
Hb_121467_010 |
0.0971466987 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 3 |
Hb_142751_010 |
0.1505133234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_178041_010 |
0.1505171347 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 5 |
Hb_130134_030 |
0.1505852771 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 6 |
Hb_009119_030 |
0.1603826843 |
- |
- |
GMFP4 [Medicago truncatula] |
| 7 |
Hb_000486_060 |
0.1616854982 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 8 |
Hb_001767_120 |
0.166755138 |
- |
- |
- |
| 9 |
Hb_004935_080 |
0.180607362 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 10 |
Hb_070469_010 |
0.1820144163 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 11 |
Hb_001623_300 |
0.1824324498 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 12 |
Hb_002742_070 |
0.1831378503 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 13 |
Hb_000062_060 |
0.1862325849 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 14 |
Hb_037793_010 |
0.1865302582 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 15 |
Hb_130134_020 |
0.1868008211 |
- |
- |
- |
| 16 |
Hb_051437_010 |
0.1872950498 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002411_130 |
0.1889508012 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 18 |
Hb_000130_190 |
0.1906377438 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 19 |
Hb_010964_020 |
0.1912635074 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 20 |
Hb_070385_010 |
0.1913817624 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |