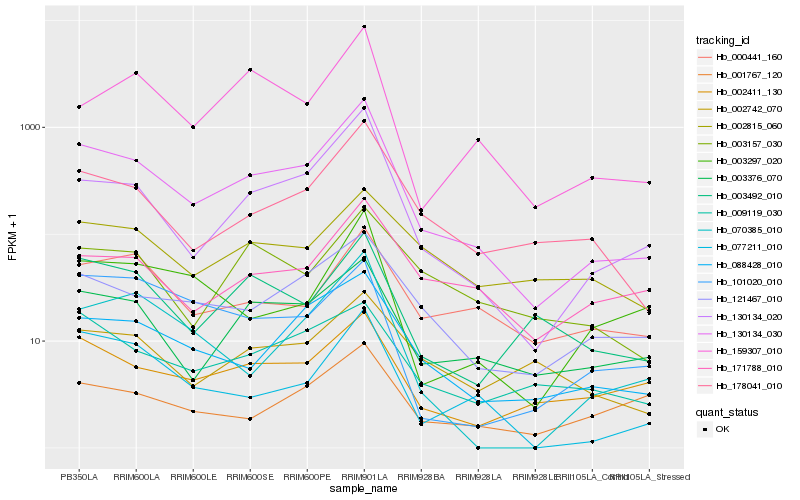
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130134_030 |
0.0 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 2 |
Hb_130134_020 |
0.1137965719 |
- |
- |
- |
| 3 |
Hb_178041_010 |
0.1457929747 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 4 |
Hb_088428_010 |
0.1505852771 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 5 |
Hb_003376_070 |
0.1508811407 |
- |
- |
hypothetical protein B456_009G419800 [Gossypium raimondii] |
| 6 |
Hb_121467_010 |
0.1607485623 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 7 |
Hb_077211_010 |
0.1675614016 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 8 |
Hb_002411_130 |
0.1751577944 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 9 |
Hb_159307_010 |
0.1778719287 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_003492_010 |
0.182201326 |
- |
- |
DREPP plasma membrane polypeptide family protein [Populus trichocarpa] |
| 11 |
Hb_009119_030 |
0.1824869488 |
- |
- |
GMFP4 [Medicago truncatula] |
| 12 |
Hb_070385_010 |
0.1857273426 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 13 |
Hb_003297_020 |
0.18835922 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 14 |
Hb_171788_010 |
0.1893072083 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 15 |
Hb_000441_160 |
0.1932174068 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_002742_070 |
0.196601243 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 17 |
Hb_003157_030 |
0.2000372843 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 18 |
Hb_001767_120 |
0.2045431523 |
- |
- |
- |
| 19 |
Hb_002815_060 |
0.2076397564 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 20 |
Hb_101020_010 |
0.208268929 |
- |
- |
- |