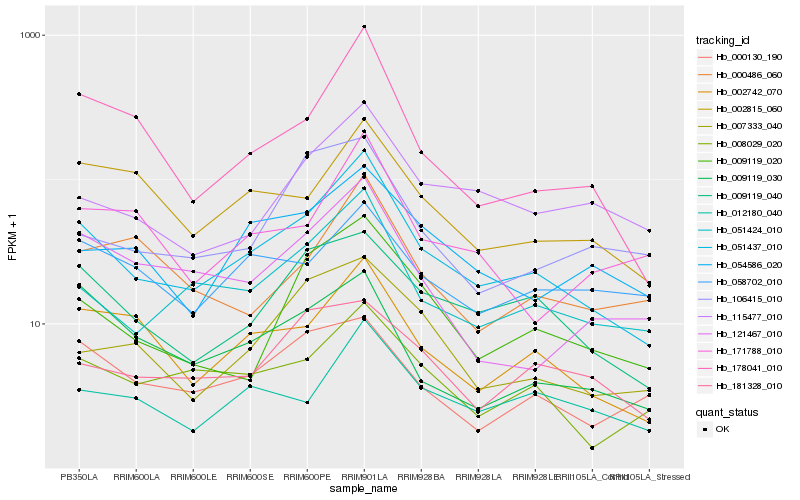
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_051437_010 |
0.0 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 2 |
Hb_051424_010 |
0.1219084305 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 3 |
Hb_002742_070 |
0.134641721 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 4 |
Hb_178041_010 |
0.1385336454 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 5 |
Hb_009119_040 |
0.1456451226 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 6 |
Hb_008029_020 |
0.1466854823 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 7 |
Hb_009119_020 |
0.1486531834 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 8 |
Hb_012180_040 |
0.1520190072 |
- |
- |
hypothetical protein VITISV_042517 [Vitis vinifera] |
| 9 |
Hb_115477_010 |
0.1595350762 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 10 |
Hb_002815_060 |
0.1602950997 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 11 |
Hb_121467_010 |
0.1619923214 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 12 |
Hb_054586_020 |
0.1627109376 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 13 |
Hb_007333_040 |
0.1650950471 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 14 |
Hb_171788_010 |
0.1719086009 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 15 |
Hb_181328_010 |
0.1731866766 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000130_190 |
0.1745419074 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 17 |
Hb_009119_030 |
0.177902766 |
- |
- |
GMFP4 [Medicago truncatula] |
| 18 |
Hb_000486_060 |
0.1796618239 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 19 |
Hb_058702_010 |
0.1804418822 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_106415_010 |
0.1804996083 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |