| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
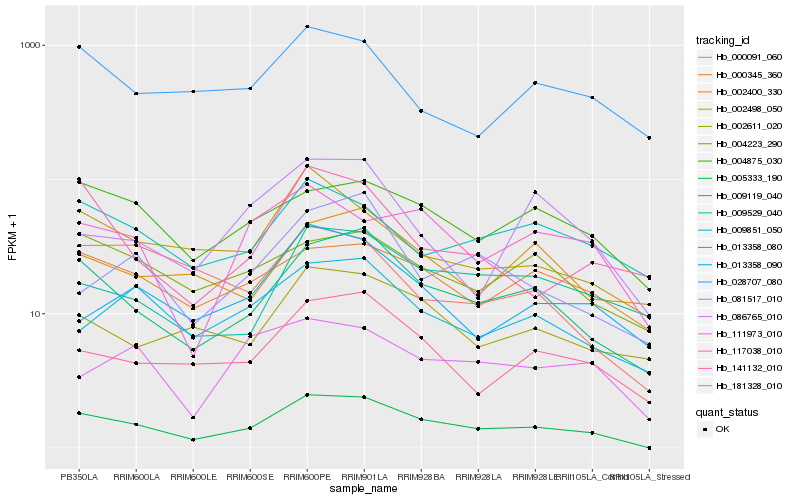
Hb_005333_190 |
0.0 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X2 [Populus euphratica] |
| 2 |
Hb_009119_040 |
0.1181898117 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 3 |
Hb_181328_010 |
0.165883536 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_004875_030 |
0.1791500555 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 5 |
Hb_002498_050 |
0.1800970002 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 6 |
Hb_013358_080 |
0.1834632509 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 7 |
Hb_000345_360 |
0.1850440845 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 8 |
Hb_002611_020 |
0.1864350144 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 9 |
Hb_081517_010 |
0.1870588752 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 10 |
Hb_117038_010 |
0.1873416803 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 11 |
Hb_009529_040 |
0.1878663917 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 12 |
Hb_004223_290 |
0.1899199983 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 13 |
Hb_013358_090 |
0.1907493302 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 14 |
Hb_009851_050 |
0.1927760456 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 15 |
Hb_028707_080 |
0.1961908039 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 16 |
Hb_141132_010 |
0.1968049035 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 17 |
Hb_111973_010 |
0.197779502 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_086765_010 |
0.1981096843 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 19 |
Hb_000091_060 |
0.2000669136 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 20 |
Hb_002400_330 |
0.2006892508 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |