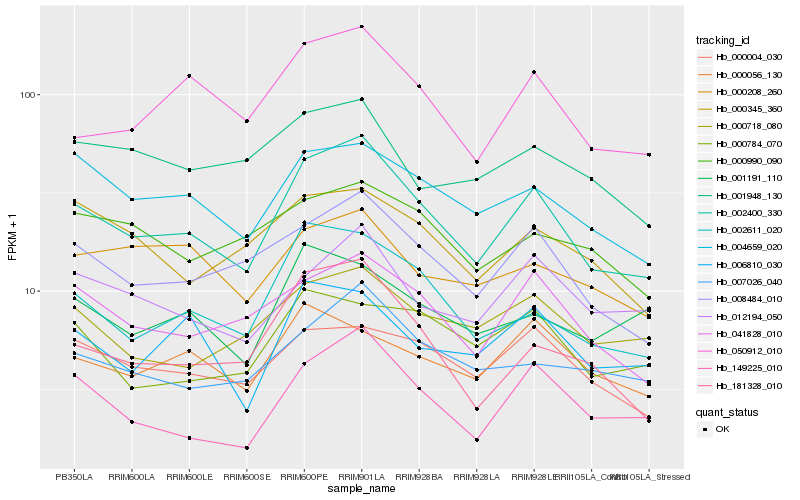
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_330 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 2 |
Hb_008484_010 |
0.0914160011 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 3 |
Hb_002611_020 |
0.1014616403 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 4 |
Hb_041828_010 |
0.1021380927 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 5 |
Hb_050912_010 |
0.1053868602 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_149225_010 |
0.1069511627 |
- |
- |
DNA-directed RNA polymerase II subunit rpb4, putative isoform 4, partial [Theobroma cacao] |
| 7 |
Hb_004659_020 |
0.1092066447 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 8 |
Hb_181328_010 |
0.1094234032 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_007026_040 |
0.113246369 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 10 |
Hb_000004_030 |
0.1157550752 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 11 |
Hb_012194_050 |
0.1177761493 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 12 |
Hb_000718_080 |
0.1223351305 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |
| 13 |
Hb_006810_030 |
0.1235141346 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000056_130 |
0.1271904607 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_000345_360 |
0.1276268828 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 16 |
Hb_000784_070 |
0.1303156091 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001191_110 |
0.1304075838 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 18 |
Hb_001948_130 |
0.1320892622 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 19 |
Hb_000990_090 |
0.1321715686 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_000208_260 |
0.1322359562 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |