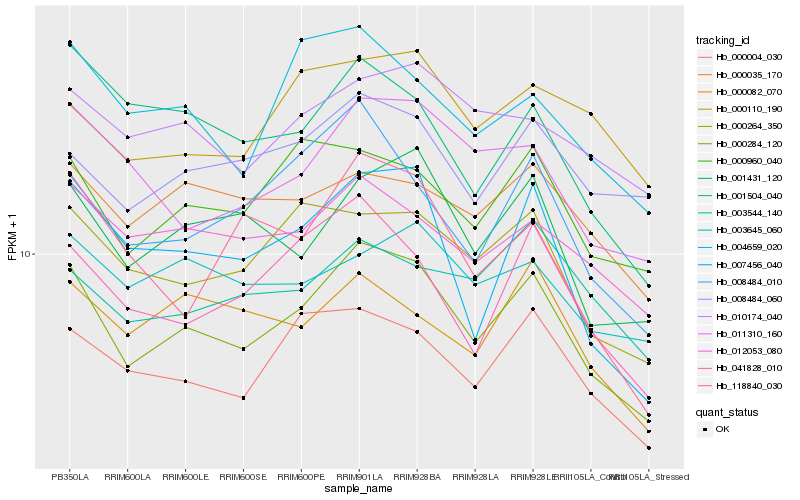
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |
| 2 |
Hb_000004_030 |
0.0860124684 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 3 |
Hb_001431_120 |
0.0898798954 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 4 |
Hb_041828_010 |
0.0903850061 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 5 |
Hb_000264_350 |
0.09082002 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 6 |
Hb_007456_040 |
0.0923560631 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 7 |
Hb_000960_040 |
0.0930013237 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 8 |
Hb_008484_010 |
0.0975840146 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 9 |
Hb_011310_160 |
0.0976931927 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 10 |
Hb_012053_080 |
0.1002729451 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 11 |
Hb_004659_020 |
0.1027846986 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 12 |
Hb_008484_060 |
0.1053478782 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 13 |
Hb_010174_040 |
0.1077212818 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 14 |
Hb_000035_170 |
0.1087580016 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 15 |
Hb_000082_070 |
0.1093324359 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 16 |
Hb_001504_040 |
0.1108953384 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 17 |
Hb_000110_190 |
0.1110167328 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 18 |
Hb_003645_060 |
0.1111058847 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_118840_030 |
0.1116412111 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 20 |
Hb_003544_140 |
0.1129736813 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |