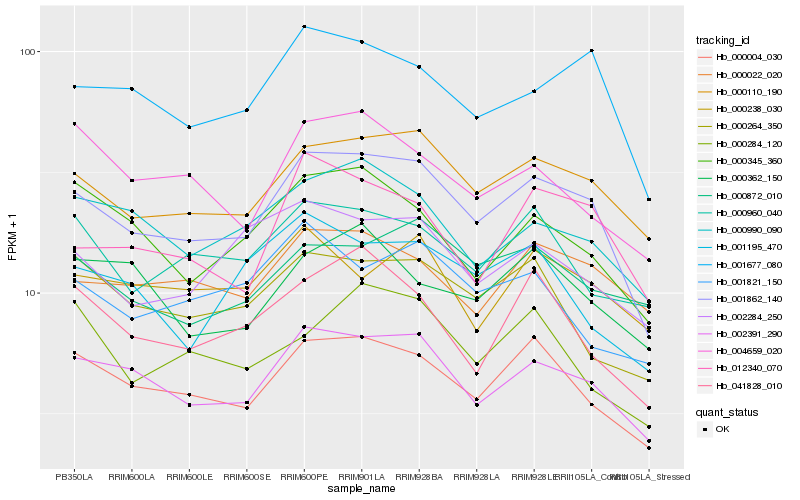
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_140 |
0.0 |
- |
- |
natural resistance-associated macrophage protein, putative [Ricinus communis] |
| 2 |
Hb_002391_290 |
0.0640974546 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000110_190 |
0.0712983923 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 4 |
Hb_000004_030 |
0.0760378101 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 5 |
Hb_001677_080 |
0.082460822 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000990_090 |
0.0985843596 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 7 |
Hb_012340_070 |
0.0990167845 |
- |
- |
Gamma-secretase subunit APH1-like family protein [Populus trichocarpa] |
| 8 |
Hb_000345_360 |
0.1028511937 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 9 |
Hb_000264_350 |
0.1046576203 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 10 |
Hb_000022_020 |
0.1050253258 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 11 |
Hb_041828_010 |
0.1058720973 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 12 |
Hb_002284_250 |
0.1059062031 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 13 |
Hb_000960_040 |
0.1073802438 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 14 |
Hb_004659_020 |
0.1093705757 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 15 |
Hb_000872_010 |
0.1102253775 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 16 |
Hb_001195_470 |
0.1106079554 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 17 |
Hb_000238_030 |
0.1139158962 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001821_150 |
0.1144679418 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 19 |
Hb_000362_150 |
0.1149526789 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 20 |
Hb_000284_120 |
0.1154771613 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |