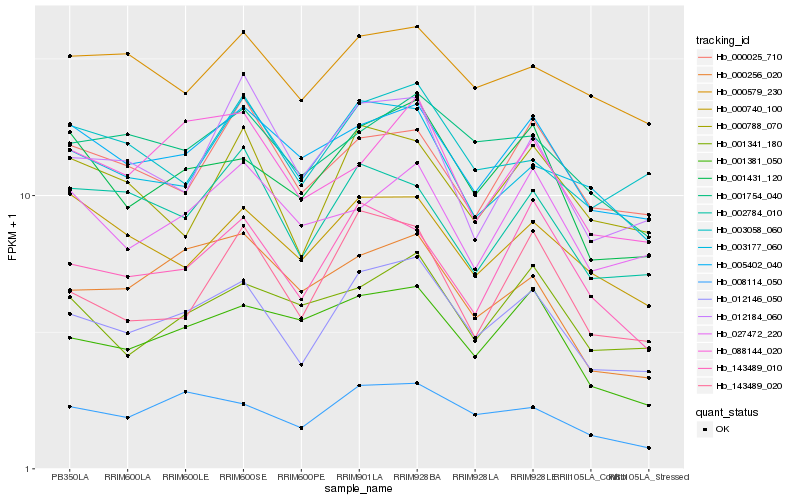
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012146_050 |
0.0 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001431_120 |
0.077167517 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 3 |
Hb_008114_050 |
0.0826741975 |
- |
- |
hypothetical protein POPTR_0006s02430g [Populus trichocarpa] |
| 4 |
Hb_005402_040 |
0.0873527009 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 5 |
Hb_143489_010 |
0.0924375385 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 6 |
Hb_143489_020 |
0.0947565382 |
- |
- |
- |
| 7 |
Hb_000788_070 |
0.0957879309 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 8 |
Hb_001754_040 |
0.0958222413 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 9 |
Hb_088144_020 |
0.0967043199 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_003177_060 |
0.0991148923 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003058_060 |
0.0994403048 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 12 |
Hb_001381_050 |
0.099560812 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_002784_010 |
0.0998968587 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 14 |
Hb_001341_180 |
0.1018951194 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 15 |
Hb_000256_020 |
0.1019967721 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 16 |
Hb_012184_060 |
0.1035356865 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000025_710 |
0.1040785196 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 18 |
Hb_000740_100 |
0.1041317543 |
- |
- |
calpain, putative [Ricinus communis] |
| 19 |
Hb_027472_220 |
0.104521724 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000579_230 |
0.1051304037 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |