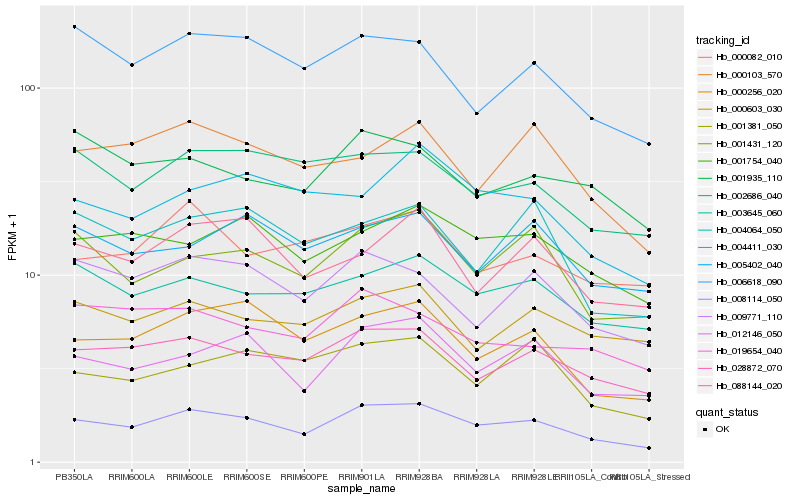
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008114_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s02430g [Populus trichocarpa] |
| 2 |
Hb_012146_050 |
0.0826741975 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_028872_070 |
0.0832573602 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000256_020 |
0.0902896541 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 5 |
Hb_001431_120 |
0.0935052746 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 6 |
Hb_003645_060 |
0.0973160157 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001754_040 |
0.097833312 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 8 |
Hb_009771_110 |
0.0978549699 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 9 |
Hb_006618_090 |
0.1033118814 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 10 |
Hb_002686_040 |
0.1037746338 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 11 |
Hb_088144_020 |
0.1072323747 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000103_570 |
0.1085568377 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001381_050 |
0.1088395027 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 14 |
Hb_004411_030 |
0.10910375 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 15 |
Hb_000603_030 |
0.1091757051 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 16 |
Hb_001935_110 |
0.1099904495 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 17 |
Hb_004064_050 |
0.1101212782 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 18 |
Hb_005402_040 |
0.1107915371 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 19 |
Hb_000082_010 |
0.1109385837 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 20 |
Hb_019654_040 |
0.1114098571 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |