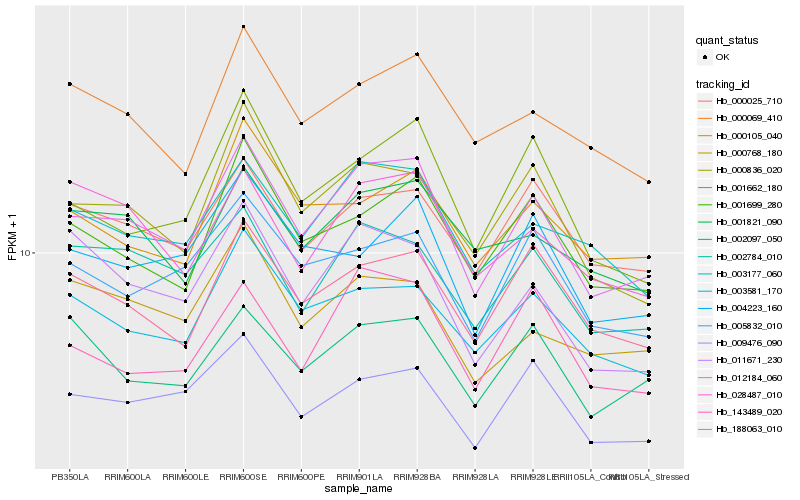
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012184_060 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000836_020 |
0.0765160009 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 3 |
Hb_003177_060 |
0.0774581779 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009476_090 |
0.0777397765 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_001699_280 |
0.0794564321 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 6 |
Hb_002784_010 |
0.0797759237 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 7 |
Hb_188063_010 |
0.0798478777 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 8 |
Hb_000025_710 |
0.0824760899 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 9 |
Hb_003581_170 |
0.0828178758 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005832_010 |
0.0836747064 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001662_180 |
0.0841082545 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 12 |
Hb_143489_020 |
0.0885348391 |
- |
- |
- |
| 13 |
Hb_000069_410 |
0.0888872207 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 14 |
Hb_002097_050 |
0.0893399068 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 15 |
Hb_001821_090 |
0.0915081746 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 16 |
Hb_004223_160 |
0.0918048725 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011671_230 |
0.0918566691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000105_040 |
0.0929105131 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 19 |
Hb_000768_180 |
0.0931650559 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 20 |
Hb_028487_010 |
0.0933396146 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |