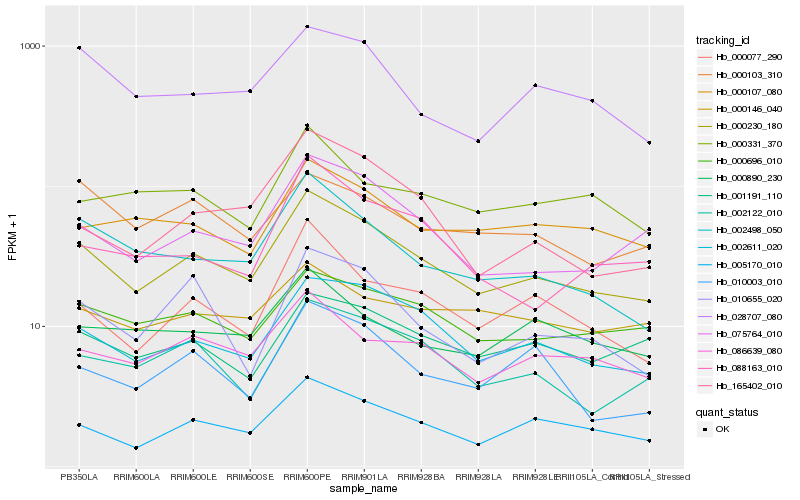
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_180 |
0.0 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_002611_020 |
0.0856289799 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 3 |
Hb_005170_010 |
0.0901884102 |
- |
- |
- |
| 4 |
Hb_075764_010 |
0.1000461489 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 5 |
Hb_086639_080 |
0.1044886134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002498_050 |
0.1052804533 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 7 |
Hb_000696_010 |
0.1068768236 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 8 |
Hb_002122_010 |
0.10930716 |
- |
- |
- |
| 9 |
Hb_000077_290 |
0.1108152298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_028707_080 |
0.1166423326 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 11 |
Hb_001191_110 |
0.118219089 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 12 |
Hb_000107_080 |
0.1197235935 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 13 |
Hb_000890_230 |
0.1246954471 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 14 |
Hb_010655_020 |
0.1249437281 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 15 |
Hb_165402_010 |
0.1262226321 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 16 |
Hb_010003_010 |
0.12678766 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000103_310 |
0.1271687821 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 18 |
Hb_000331_370 |
0.1290776626 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 19 |
Hb_000146_040 |
0.1308117484 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 20 |
Hb_088163_010 |
0.1309466514 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |