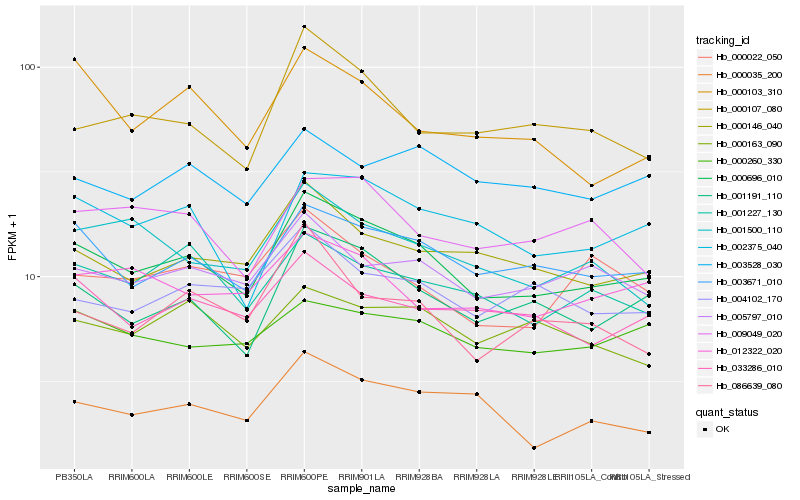
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_010 |
0.0 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 2 |
Hb_001191_110 |
0.069966427 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 3 |
Hb_003671_010 |
0.0710408812 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000146_040 |
0.0872253604 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 5 |
Hb_002375_040 |
0.0890186441 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 6 |
Hb_005797_010 |
0.0910777716 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_033286_010 |
0.0959114463 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 8 |
Hb_001227_130 |
0.0969822942 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 9 |
Hb_000107_080 |
0.0994891866 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 10 |
Hb_001500_110 |
0.0996509114 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_004102_170 |
0.0996968128 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 12 |
Hb_012322_020 |
0.101619504 |
- |
- |
- |
| 13 |
Hb_000260_330 |
0.1016834175 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |
| 14 |
Hb_000035_200 |
0.102886103 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |
| 15 |
Hb_003528_030 |
0.1030896186 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 16 |
Hb_086639_080 |
0.104390929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009049_020 |
0.1055364282 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 18 |
Hb_000163_090 |
0.105615315 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 19 |
Hb_000022_050 |
0.1061497932 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 20 |
Hb_000103_310 |
0.1061958659 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |