| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
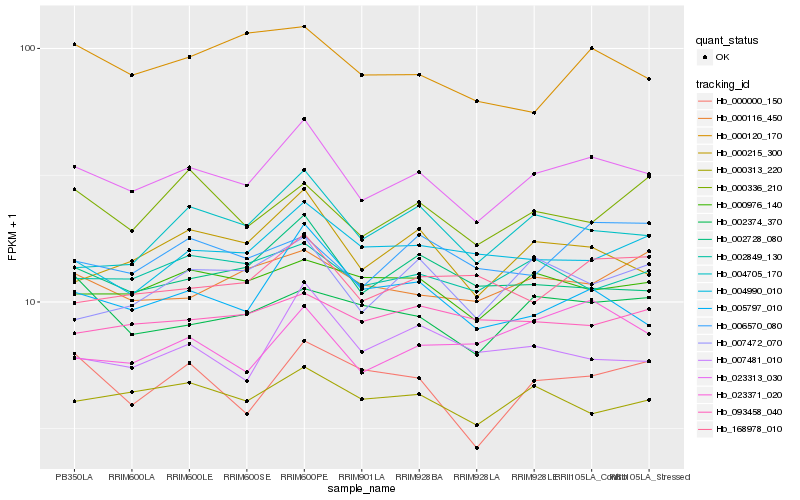
Hb_023313_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002728_080 |
0.0691233433 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_005797_010 |
0.0700477305 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000215_300 |
0.0704375674 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 5 |
Hb_000313_220 |
0.0719534096 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000976_140 |
0.0723751311 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002849_130 |
0.0749234076 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 8 |
Hb_004705_170 |
0.0770744371 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 9 |
Hb_002374_370 |
0.0808721503 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_004990_010 |
0.0810801549 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_000116_450 |
0.0817964746 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_007472_070 |
0.0825418557 |
- |
- |
cir, putative [Ricinus communis] |
| 13 |
Hb_168978_010 |
0.0830269073 |
- |
- |
PREDICTED: uncharacterized protein LOC105630751 [Jatropha curcas] |
| 14 |
Hb_000000_150 |
0.0844437917 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 15 |
Hb_023371_020 |
0.0845813872 |
- |
- |
PREDICTED: uncharacterized protein LOC105633512 [Jatropha curcas] |
| 16 |
Hb_000336_210 |
0.0848472785 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000120_170 |
0.0851398719 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 18 |
Hb_007481_010 |
0.0852370475 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 19 |
Hb_093458_040 |
0.0860383088 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 20 |
Hb_006570_080 |
0.0865149906 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |