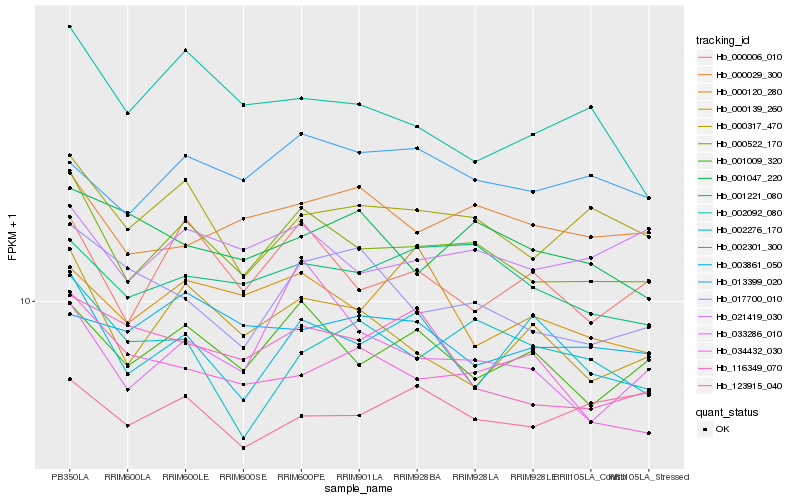
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000522_170 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 2 |
Hb_000317_470 |
0.0634872769 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 3 |
Hb_001009_320 |
0.0639816461 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 4 |
Hb_000006_010 |
0.0673419451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001221_080 |
0.070561708 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_021419_030 |
0.0712318246 |
- |
- |
hypothetical protein glysoja_023295 [Glycine soja] |
| 7 |
Hb_000139_260 |
0.0759881226 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 8 |
Hb_000029_300 |
0.0794181439 |
- |
- |
hypothetical protein JCGZ_10048 [Jatropha curcas] |
| 9 |
Hb_013399_020 |
0.0800827993 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 10 |
Hb_033286_010 |
0.0810820255 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 11 |
Hb_017700_010 |
0.0812247191 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 12 |
Hb_002301_300 |
0.0832800626 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 13 |
Hb_002092_080 |
0.08552586 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 14 |
Hb_116349_070 |
0.088321501 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 15 |
Hb_000120_280 |
0.088639882 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 16 |
Hb_034432_030 |
0.0886531854 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001047_220 |
0.0886832317 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 18 |
Hb_123915_040 |
0.0903781077 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 19 |
Hb_003861_050 |
0.091229744 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 20 |
Hb_002276_170 |
0.0914016916 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |