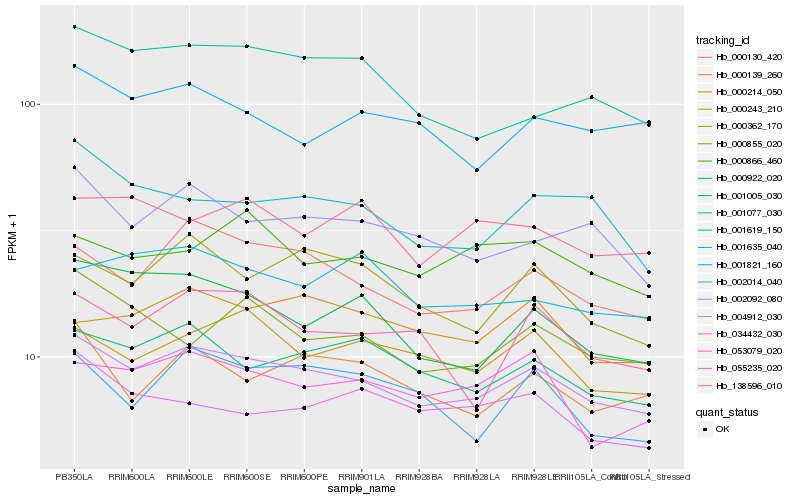
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004912_030 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 2 |
Hb_000130_420 |
0.055935863 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 3 |
Hb_001005_030 |
0.0626303158 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_002092_080 |
0.0657321125 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 5 |
Hb_000243_210 |
0.0702776686 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000362_170 |
0.0723526772 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 7 |
Hb_053079_020 |
0.0723725142 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 8 |
Hb_000139_260 |
0.0732050852 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 9 |
Hb_001619_150 |
0.0749250066 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000922_020 |
0.0750238601 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_000214_050 |
0.0762763753 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001821_160 |
0.0770291696 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 13 |
Hb_055235_020 |
0.0776968566 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002014_040 |
0.0782118879 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 15 |
Hb_001077_030 |
0.0784415809 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000855_020 |
0.078785884 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 17 |
Hb_001635_040 |
0.0788925548 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_138596_010 |
0.0791252134 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 19 |
Hb_000866_460 |
0.0793439196 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_034432_030 |
0.079528907 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |