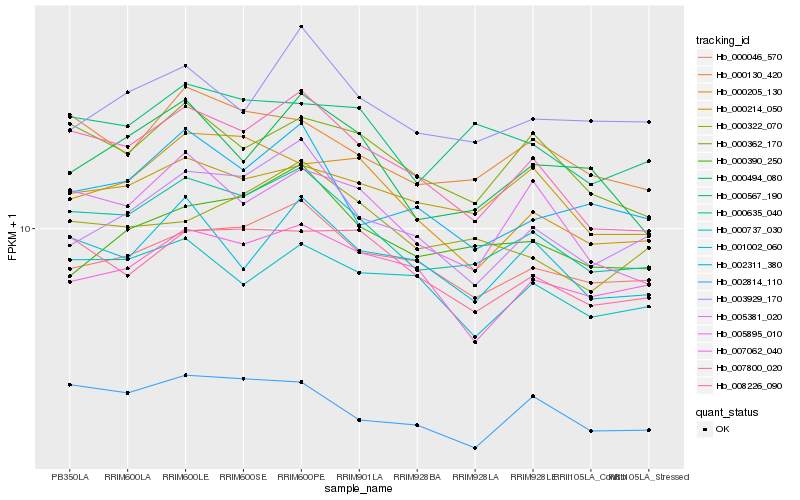
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_040 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 2 |
Hb_007800_020 |
0.0535782426 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000130_420 |
0.0694251774 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 4 |
Hb_000046_570 |
0.0706785403 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 5 |
Hb_000362_170 |
0.072511618 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_000567_190 |
0.0759219262 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_008226_090 |
0.0763432699 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005895_010 |
0.0770969931 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005381_020 |
0.0784102092 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 10 |
Hb_001002_060 |
0.080587419 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 11 |
Hb_000494_080 |
0.0818794373 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000214_050 |
0.0828110059 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002311_380 |
0.0829774996 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 14 |
Hb_002814_110 |
0.0839532322 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 15 |
Hb_000737_030 |
0.0847641984 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007062_040 |
0.0850848053 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 17 |
Hb_000390_250 |
0.0860921926 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 18 |
Hb_000205_130 |
0.0867385051 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000322_070 |
0.087062706 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_003929_170 |
0.0874660975 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |