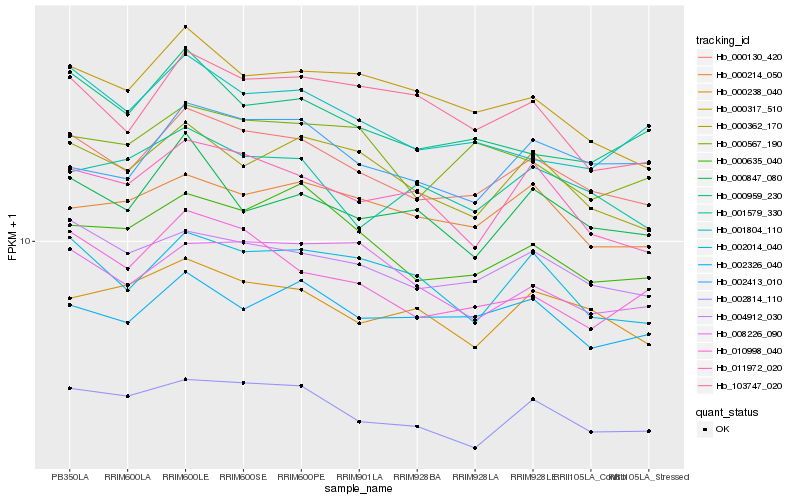
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_420 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 2 |
Hb_004912_030 |
0.055935863 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 3 |
Hb_000362_170 |
0.066811016 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 4 |
Hb_000635_040 |
0.0694251774 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 5 |
Hb_011972_020 |
0.0696312629 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_002014_040 |
0.0717568862 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 7 |
Hb_000238_040 |
0.0742023325 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 8 |
Hb_000317_510 |
0.0750022483 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 9 |
Hb_002326_040 |
0.0759277022 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 10 |
Hb_002413_010 |
0.076498495 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000959_230 |
0.0767499671 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 12 |
Hb_000567_190 |
0.0780214082 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001579_330 |
0.0782233913 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000847_080 |
0.0784227858 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_000214_050 |
0.0785402861 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010998_040 |
0.0786981829 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 17 |
Hb_103747_020 |
0.0792476142 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 18 |
Hb_001804_110 |
0.0793753539 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 19 |
Hb_002814_110 |
0.079876611 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 20 |
Hb_008226_090 |
0.0808147927 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |