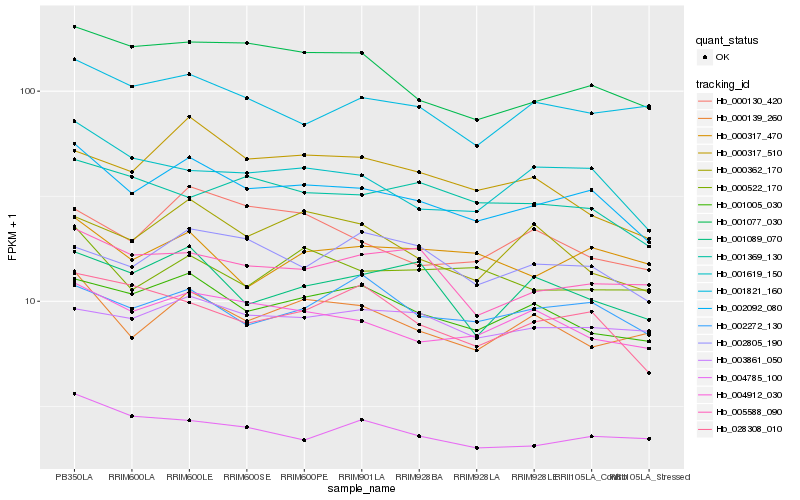
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002092_080 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 2 |
Hb_004912_030 |
0.0657321125 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 3 |
Hb_001005_030 |
0.0706010031 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_001619_150 |
0.0720464035 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001077_030 |
0.0765572856 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000139_260 |
0.0771658086 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 7 |
Hb_004785_100 |
0.0791743752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_028308_010 |
0.0811006717 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001821_160 |
0.0820036577 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 10 |
Hb_005588_090 |
0.0828639315 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 11 |
Hb_003861_050 |
0.0837795441 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 12 |
Hb_002272_130 |
0.0843261456 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 13 |
Hb_000522_170 |
0.08552586 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 14 |
Hb_000130_420 |
0.0863599115 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_001089_070 |
0.0866698748 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_001369_130 |
0.0871745874 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002805_190 |
0.0880107023 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 18 |
Hb_000317_470 |
0.088690123 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 19 |
Hb_000362_170 |
0.0890339981 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 20 |
Hb_000317_510 |
0.0905724712 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |