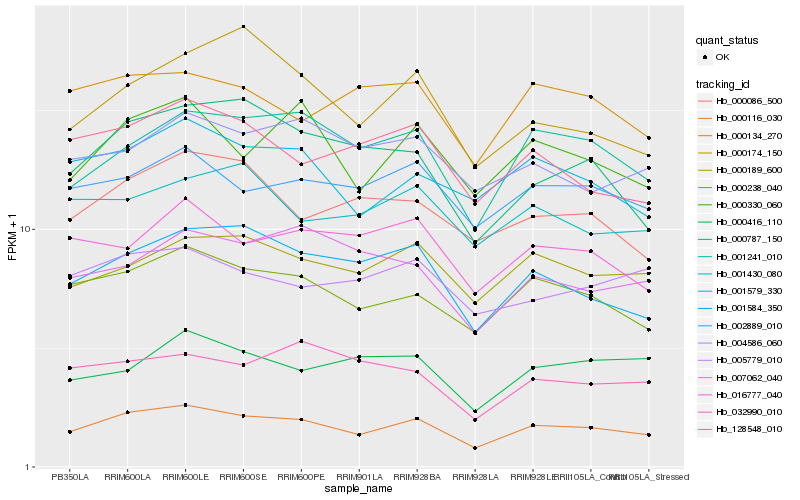
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_030 |
0.0 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 2 |
Hb_000238_040 |
0.0642927966 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 3 |
Hb_001241_010 |
0.0761829872 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001584_350 |
0.0765839724 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 5 |
Hb_000330_060 |
0.0799514051 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 6 |
Hb_001579_330 |
0.0803167484 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002889_010 |
0.0822002842 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 8 |
Hb_000189_600 |
0.0827006401 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_128548_010 |
0.0905421094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000787_150 |
0.0908038818 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 11 |
Hb_000086_500 |
0.0909562124 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_016777_040 |
0.0933448288 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 13 |
Hb_004586_060 |
0.094480379 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 14 |
Hb_000134_270 |
0.094851186 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 15 |
Hb_007062_040 |
0.0957297444 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 16 |
Hb_001430_080 |
0.0959240307 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 17 |
Hb_000174_150 |
0.0975768649 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 18 |
Hb_005779_010 |
0.0979652135 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_000416_110 |
0.0989912623 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_032990_010 |
0.0993391041 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |