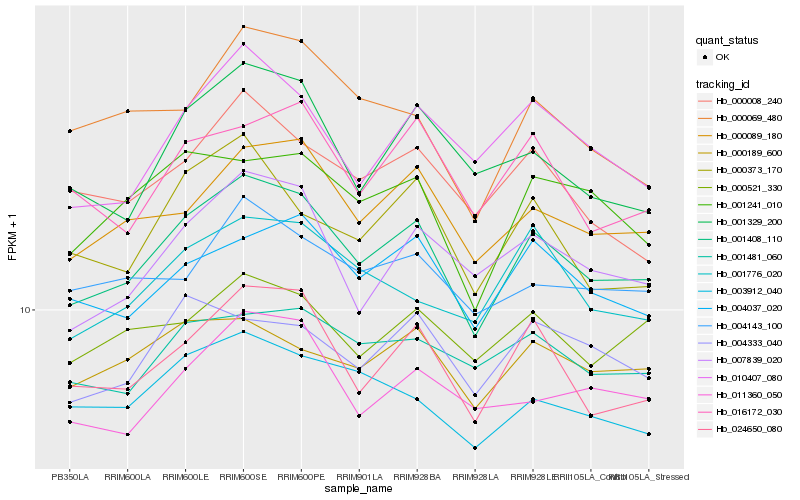
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_110 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 2 |
Hb_024650_080 |
0.058391544 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 3 |
Hb_000089_180 |
0.0609543131 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 4 |
Hb_004037_020 |
0.0692008041 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007839_020 |
0.0707568623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001241_010 |
0.072091316 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_010407_080 |
0.0760103764 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003912_040 |
0.0760136444 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 9 |
Hb_000373_170 |
0.0770322707 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 10 |
Hb_016172_030 |
0.0772788559 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 11 |
Hb_001329_200 |
0.0779317016 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 12 |
Hb_011360_050 |
0.0781994948 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000008_240 |
0.0783797047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001776_020 |
0.079096259 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 15 |
Hb_000189_600 |
0.07973107 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_001481_060 |
0.0797527478 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000521_330 |
0.079761751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004143_100 |
0.0809337556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004333_040 |
0.0810107508 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 20 |
Hb_000069_480 |
0.081157696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |