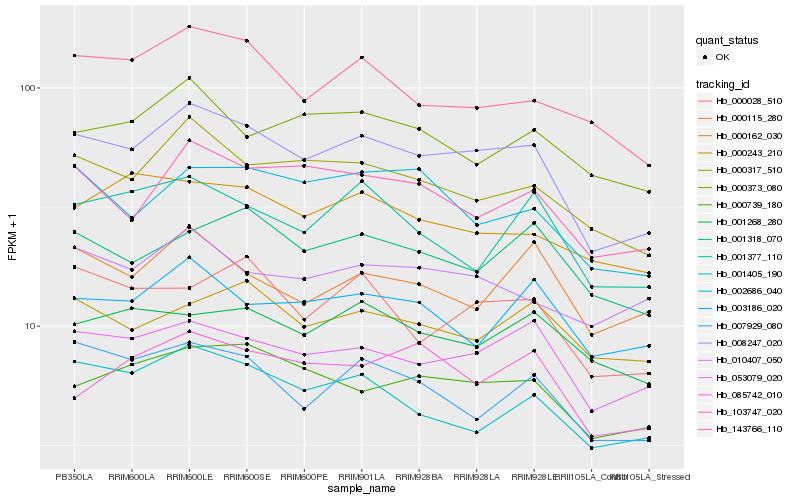
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008247_020 |
0.0 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |
| 2 |
Hb_000317_510 |
0.0692554081 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 3 |
Hb_053079_020 |
0.0697951558 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 4 |
Hb_000739_180 |
0.07466197 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 5 |
Hb_103747_020 |
0.0768542411 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 6 |
Hb_007929_080 |
0.0827800406 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 7 |
Hb_001405_190 |
0.0828924157 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_143766_110 |
0.0830299748 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 9 |
Hb_001318_070 |
0.0841241936 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002686_040 |
0.084193023 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 11 |
Hb_000028_510 |
0.0862050279 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 12 |
Hb_010407_050 |
0.0886112803 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 13 |
Hb_003186_020 |
0.0888212215 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001377_110 |
0.0892905605 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 15 |
Hb_000162_030 |
0.0898048925 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000243_210 |
0.0898799495 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_085742_010 |
0.0901646424 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |
| 18 |
Hb_000373_080 |
0.0903850732 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 19 |
Hb_001268_280 |
0.0912836326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000115_280 |
0.0913931324 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |