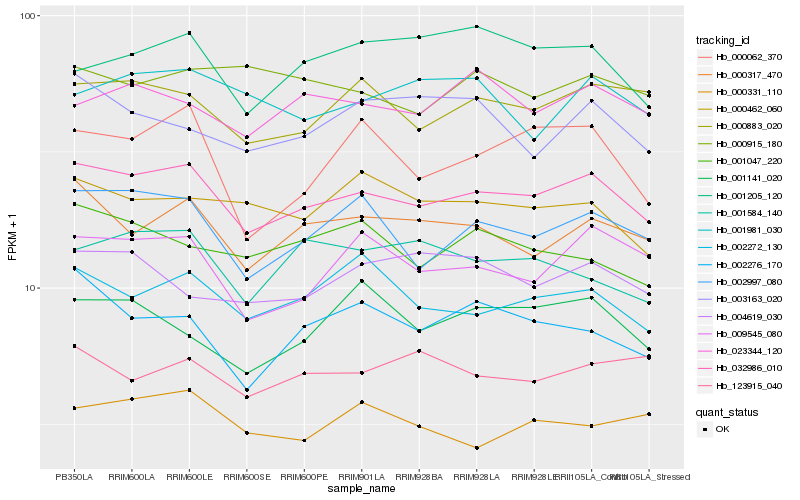
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032986_010 |
0.0 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 2 |
Hb_002997_080 |
0.0574557164 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 3 |
Hb_000317_470 |
0.060430128 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 4 |
Hb_000883_020 |
0.0621491299 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 5 |
Hb_009545_080 |
0.0668111741 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 6 |
Hb_002272_130 |
0.0688823425 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 7 |
Hb_023344_120 |
0.0736182733 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000462_060 |
0.0744292773 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001047_220 |
0.0761513555 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 10 |
Hb_000062_370 |
0.0781065166 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 11 |
Hb_002276_170 |
0.0784212053 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001584_140 |
0.0788673493 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 13 |
Hb_004619_030 |
0.0795731258 |
- |
- |
PREDICTED: uncharacterized protein LOC105633364 isoform X1 [Jatropha curcas] |
| 14 |
Hb_123915_040 |
0.0799482571 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 15 |
Hb_001981_030 |
0.0802764413 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_003163_020 |
0.0803240138 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000331_110 |
0.0808377176 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 18 |
Hb_000915_180 |
0.0810269931 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |
| 19 |
Hb_001141_020 |
0.0810860223 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 20 |
Hb_001205_120 |
0.0815811142 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |