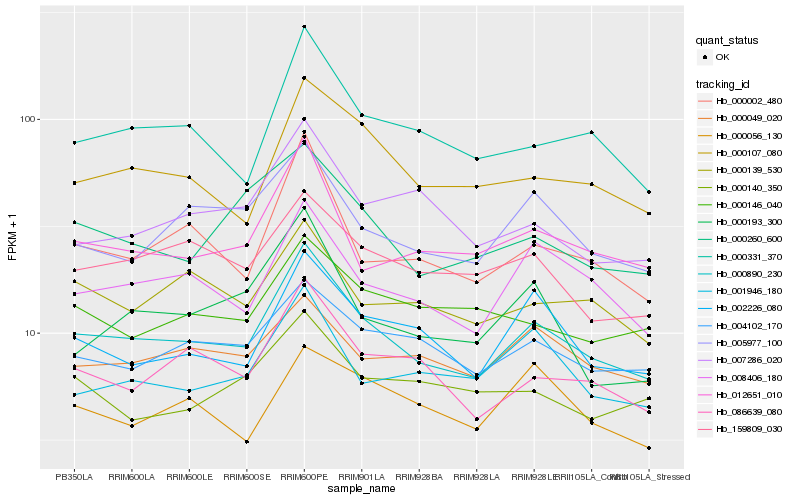
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_230 |
0.0 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 2 |
Hb_008406_180 |
0.0804844643 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_086639_080 |
0.0843665179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000107_080 |
0.0860545856 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 5 |
Hb_000331_370 |
0.0870145415 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 6 |
Hb_000002_480 |
0.0875865291 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012651_010 |
0.0893806008 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 8 |
Hb_002226_080 |
0.0902508761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005977_100 |
0.0911038862 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 10 |
Hb_004102_170 |
0.0924333516 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 11 |
Hb_159809_030 |
0.0945017828 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000139_530 |
0.0965707953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000260_600 |
0.0989236015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000049_020 |
0.1011498205 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001946_180 |
0.1025023489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007286_020 |
0.104701918 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 17 |
Hb_000146_040 |
0.106998406 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 18 |
Hb_000056_130 |
0.1116161278 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 19 |
Hb_000140_350 |
0.1124559702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000193_300 |
0.1128131395 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |