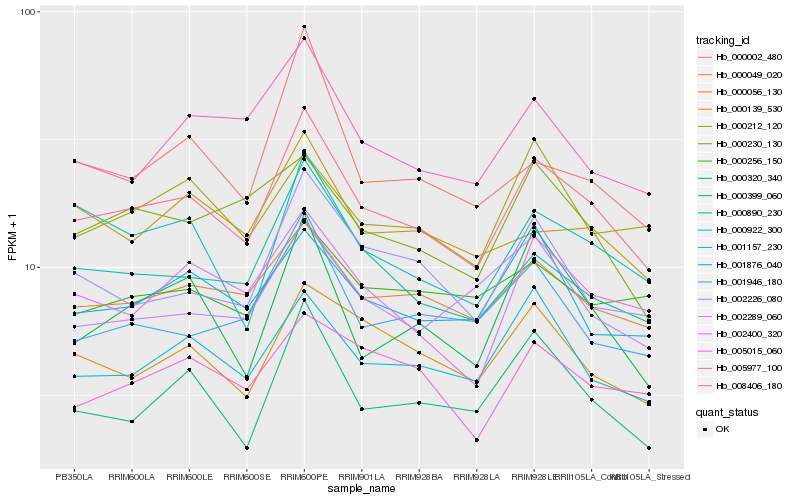
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_180 |
0.0 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_000049_020 |
0.0789924556 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000890_230 |
0.0804844643 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 4 |
Hb_002400_320 |
0.0871903031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001157_230 |
0.087651845 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_000056_130 |
0.089663242 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 7 |
Hb_002226_080 |
0.0904829368 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000922_300 |
0.0920814571 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g62930 isoform X1 [Cucumis melo] |
| 9 |
Hb_005977_100 |
0.0927218542 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 10 |
Hb_001946_180 |
0.0969211828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005015_060 |
0.0971373638 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 12 |
Hb_001876_040 |
0.0978294729 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 13 |
Hb_000399_060 |
0.0987599736 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 14 |
Hb_000230_130 |
0.1004733363 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000139_530 |
0.1011511871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000212_120 |
0.1029113199 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 17 |
Hb_000256_150 |
0.1036808757 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 18 |
Hb_000320_340 |
0.104284439 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 19 |
Hb_000002_480 |
0.104495379 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002289_060 |
0.1050988193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |