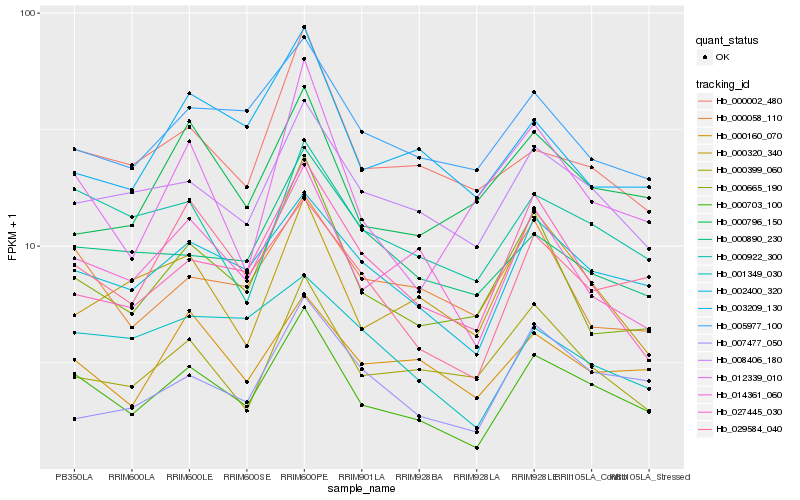
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_100 |
0.0 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase-like [Jatropha curcas] |
| 2 |
Hb_002400_320 |
0.0986680749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_014361_060 |
0.1154004279 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 4 |
Hb_008406_180 |
0.1174934503 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_000922_300 |
0.1219735907 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g62930 isoform X1 [Cucumis melo] |
| 6 |
Hb_000665_190 |
0.1220710262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_029584_040 |
0.1242813573 |
- |
- |
hypothetical protein Csa_1G586790 [Cucumis sativus] |
| 8 |
Hb_000002_480 |
0.1310436059 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000160_070 |
0.1320501219 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 10 |
Hb_027445_030 |
0.1353678535 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_005977_100 |
0.1360263876 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 12 |
Hb_007477_050 |
0.13922711 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_012339_010 |
0.1393850358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000058_110 |
0.1395606892 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000890_230 |
0.1401309046 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 16 |
Hb_000399_060 |
0.1408316263 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 17 |
Hb_003209_130 |
0.1413204354 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 18 |
Hb_001349_030 |
0.1449475988 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000796_150 |
0.147048147 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 20 |
Hb_000320_340 |
0.1476013831 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |