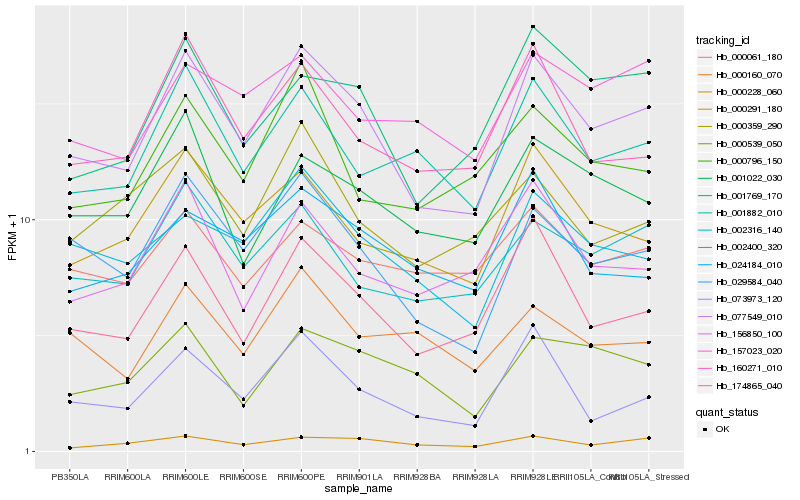
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077549_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 2 |
Hb_029584_040 |
0.0845636833 |
- |
- |
hypothetical protein Csa_1G586790 [Cucumis sativus] |
| 3 |
Hb_174865_040 |
0.0957789013 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000291_180 |
0.1023678639 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 5 |
Hb_002400_320 |
0.1052179682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002316_140 |
0.1081028275 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 7 |
Hb_160271_010 |
0.1103036406 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001882_010 |
0.1105389877 |
- |
- |
- |
| 9 |
Hb_001769_170 |
0.1114085777 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 10 |
Hb_001022_030 |
0.1169177721 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 11 |
Hb_000228_060 |
0.1204244215 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 12 |
Hb_000539_050 |
0.1209370082 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 13 |
Hb_073973_120 |
0.1210667336 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_156850_100 |
0.1213194752 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 15 |
Hb_024184_010 |
0.121780163 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_000796_150 |
0.1231848493 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 17 |
Hb_000160_070 |
0.1265364573 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 18 |
Hb_000061_180 |
0.1268601702 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 19 |
Hb_000359_290 |
0.12760221 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_157023_020 |
0.1281060844 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |