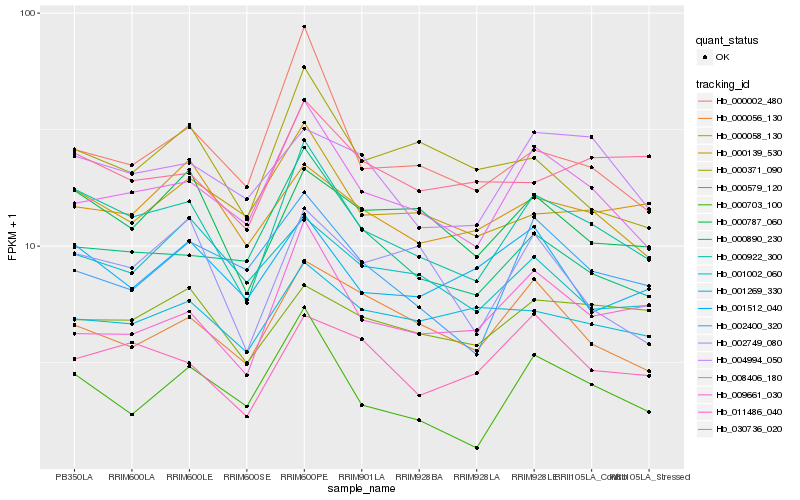
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_300 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g62930 isoform X1 [Cucumis melo] |
| 2 |
Hb_008406_180 |
0.0920814571 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_000787_060 |
0.1047517991 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000139_530 |
0.1108602513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000890_230 |
0.1173689424 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 6 |
Hb_004994_050 |
0.1183396678 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 7 |
Hb_011486_040 |
0.1186192597 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 8 |
Hb_030736_020 |
0.1210829054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000703_100 |
0.1219735907 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase-like [Jatropha curcas] |
| 10 |
Hb_000056_130 |
0.122177028 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 11 |
Hb_000002_480 |
0.12273315 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002400_320 |
0.1227403252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000579_120 |
0.1242805595 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 14 |
Hb_001512_040 |
0.1246742506 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 15 |
Hb_002749_080 |
0.1250764847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009661_030 |
0.125225086 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_000371_090 |
0.1256213953 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 18 |
Hb_001002_060 |
0.1266651298 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 19 |
Hb_001269_330 |
0.1269750666 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_000058_130 |
0.1273502949 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |