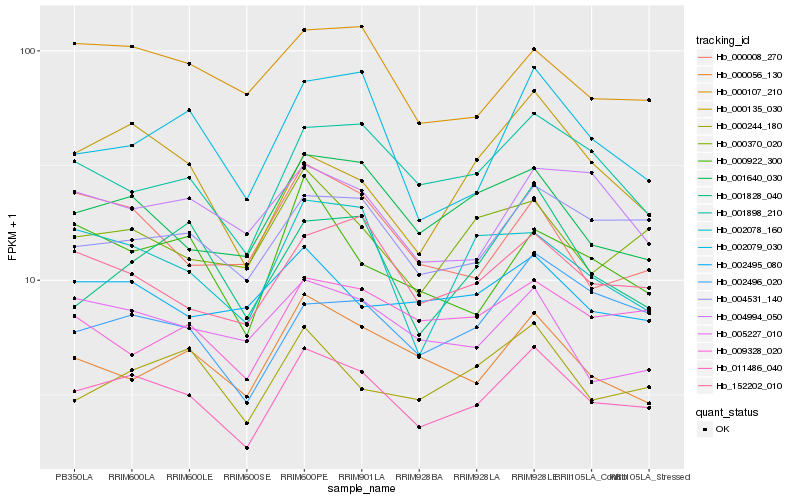
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011486_040 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 2 |
Hb_004531_140 |
0.0923854213 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 3 |
Hb_152202_010 |
0.0975684764 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 4 |
Hb_001898_210 |
0.0975702662 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 5 |
Hb_001640_030 |
0.0998669757 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 6 |
Hb_002496_020 |
0.1046655026 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000370_020 |
0.1059133462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002079_030 |
0.1074931306 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000244_180 |
0.1088030204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000107_210 |
0.1094769656 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_000056_130 |
0.1137125698 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 12 |
Hb_002078_160 |
0.1146473743 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 13 |
Hb_000135_030 |
0.1152722255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004994_050 |
0.1174654448 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 15 |
Hb_005227_010 |
0.1175815295 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001828_040 |
0.1177124338 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 17 |
Hb_000008_270 |
0.1180433422 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein REVOLUTA [Jatropha curcas] |
| 18 |
Hb_009328_020 |
0.1180496263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002495_080 |
0.1186001789 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 20 |
Hb_000922_300 |
0.1186192597 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g62930 isoform X1 [Cucumis melo] |