| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
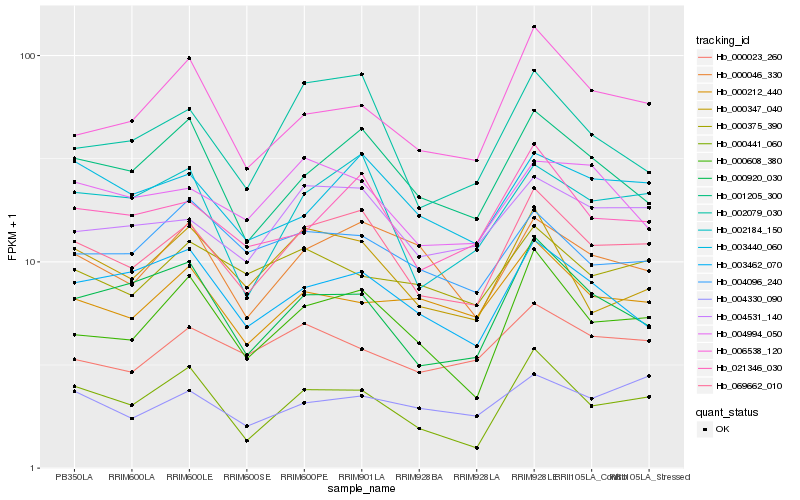
Hb_069662_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 2 |
Hb_004531_140 |
0.0820355475 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 3 |
Hb_000608_380 |
0.0821192795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021346_030 |
0.0840073735 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 5 |
Hb_002184_150 |
0.0899323298 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001205_300 |
0.0929657062 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 7 |
Hb_002079_030 |
0.0935130217 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003440_060 |
0.0989891269 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 9 |
Hb_000347_040 |
0.0996191181 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 10 |
Hb_000046_330 |
0.1012887525 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 11 |
Hb_000920_030 |
0.1064443415 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000441_060 |
0.1068156984 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 13 |
Hb_006538_120 |
0.1070129467 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003462_070 |
0.1072220785 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 15 |
Hb_000023_260 |
0.1088902477 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000375_390 |
0.1102674535 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 17 |
Hb_000212_440 |
0.1105502203 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_004994_050 |
0.1109164174 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 19 |
Hb_004096_240 |
0.1119091269 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 20 |
Hb_004330_090 |
0.1131294303 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |