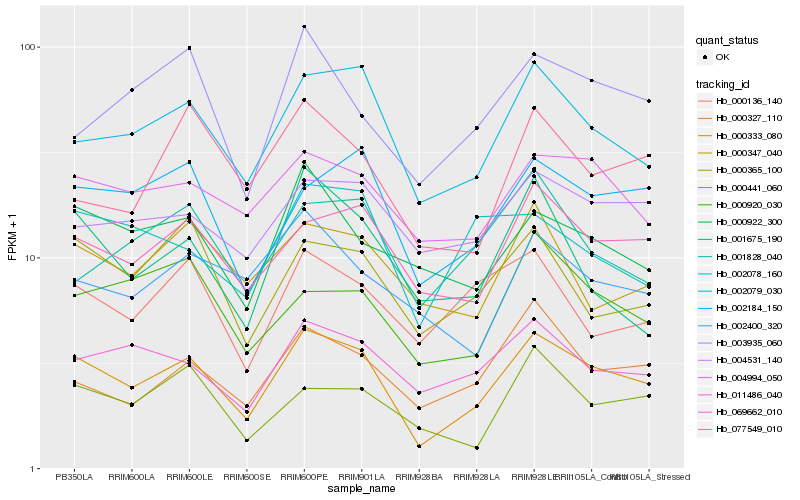
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000333_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002079_030 |
0.1099726651 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 3 |
Hb_069662_010 |
0.1223466815 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 4 |
Hb_002184_150 |
0.1255967734 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_004994_050 |
0.1338453994 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 6 |
Hb_011486_040 |
0.1347862735 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 7 |
Hb_000327_110 |
0.1394841255 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 8 |
Hb_000365_100 |
0.1408102252 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000920_030 |
0.1442915376 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004531_140 |
0.1444632302 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 11 |
Hb_000922_300 |
0.145740714 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g62930 isoform X1 [Cucumis melo] |
| 12 |
Hb_000347_040 |
0.1472711812 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 13 |
Hb_003935_060 |
0.1477426888 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 14 |
Hb_000441_060 |
0.1522626605 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 15 |
Hb_077549_010 |
0.152352518 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000136_140 |
0.1524138952 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 17 |
Hb_001675_190 |
0.1534826574 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 18 |
Hb_002400_320 |
0.1535013696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001828_040 |
0.1538155143 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 20 |
Hb_002078_160 |
0.154672519 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |