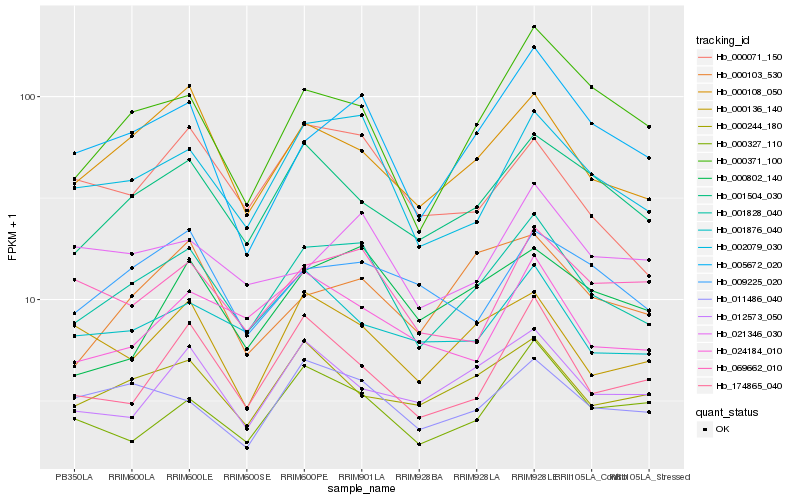
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001828_040 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 2 |
Hb_002079_030 |
0.0823782195 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000108_050 |
0.099941675 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 4 |
Hb_001504_030 |
0.1061891708 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 5 |
Hb_005672_020 |
0.1095243571 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000371_100 |
0.1170066841 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_011486_040 |
0.1177124338 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 8 |
Hb_000244_180 |
0.1218130035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_174865_040 |
0.1236914721 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_024184_010 |
0.1247777615 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_001876_040 |
0.1253230869 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 12 |
Hb_000327_110 |
0.126293828 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 13 |
Hb_000071_150 |
0.1265197835 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 14 |
Hb_000136_140 |
0.1267583096 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 15 |
Hb_069662_010 |
0.1268069276 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 16 |
Hb_012573_050 |
0.1273105562 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 17 |
Hb_000802_140 |
0.1275182337 |
- |
- |
hypothetical protein B456_011G045700 [Gossypium raimondii] |
| 18 |
Hb_000103_530 |
0.1293922858 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 19 |
Hb_009225_020 |
0.1294140244 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 20 |
Hb_021346_030 |
0.1301072352 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |