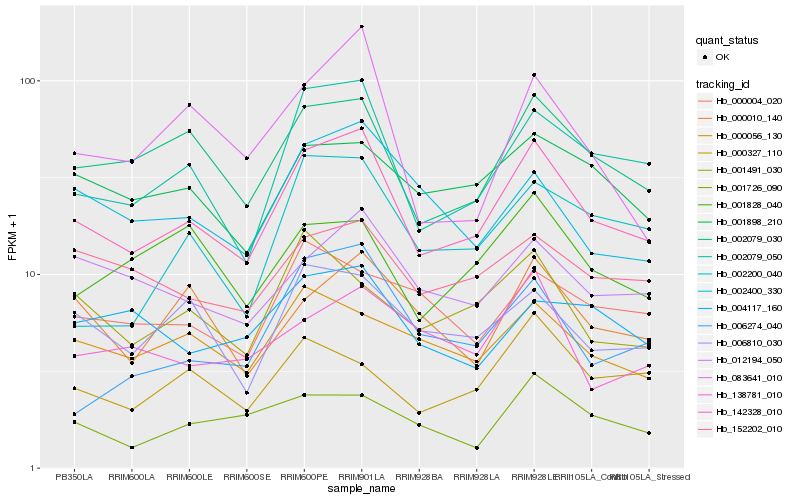
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142328_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002079_050 |
0.0903939754 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 3 |
Hb_002079_030 |
0.1030886895 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006810_030 |
0.1328883482 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_012194_050 |
0.1344315878 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 6 |
Hb_001898_210 |
0.1356250059 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 7 |
Hb_152202_010 |
0.1373726793 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 8 |
Hb_000056_130 |
0.1381456226 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 9 |
Hb_002400_330 |
0.1394657469 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_000010_140 |
0.1426164449 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000327_110 |
0.1450623313 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 12 |
Hb_001828_040 |
0.1452634687 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 13 |
Hb_083641_010 |
0.1459096971 |
- |
- |
SECY isoform 1 [Theobroma cacao] |
| 14 |
Hb_001491_030 |
0.1462478164 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002200_040 |
0.1475737977 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001726_090 |
0.1476743351 |
- |
- |
- |
| 17 |
Hb_138781_010 |
0.1484807998 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 18 |
Hb_006274_040 |
0.1488901481 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 19 |
Hb_000004_020 |
0.1506377357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004117_160 |
0.1512880721 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |