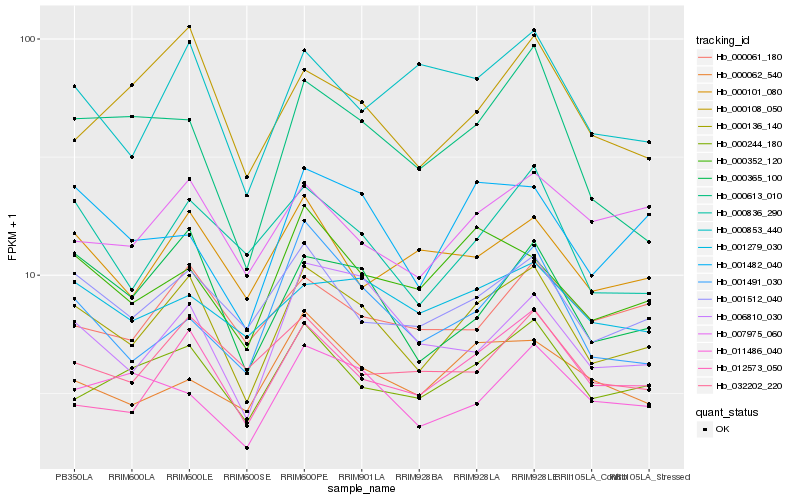
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000136_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 2 |
Hb_000365_100 |
0.0899147161 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001512_040 |
0.0974331522 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 4 |
Hb_000244_180 |
0.1015743845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012573_050 |
0.1029353995 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 6 |
Hb_000836_290 |
0.1037750694 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_006810_030 |
0.1140405843 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_000352_120 |
0.1149835329 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 9 |
Hb_001482_040 |
0.1153787461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007975_060 |
0.1177386609 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000101_080 |
0.117859377 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 12 |
Hb_000108_050 |
0.1197126454 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 13 |
Hb_011486_040 |
0.1210779006 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 14 |
Hb_000613_010 |
0.1212353294 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 15 |
Hb_032202_220 |
0.1222054679 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_001279_030 |
0.12237696 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 17 |
Hb_000853_440 |
0.1230361382 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 18 |
Hb_001491_030 |
0.1231322559 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000062_540 |
0.1236419275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000061_180 |
0.1245780075 |
- |
- |
exonuclease, putative [Ricinus communis] |