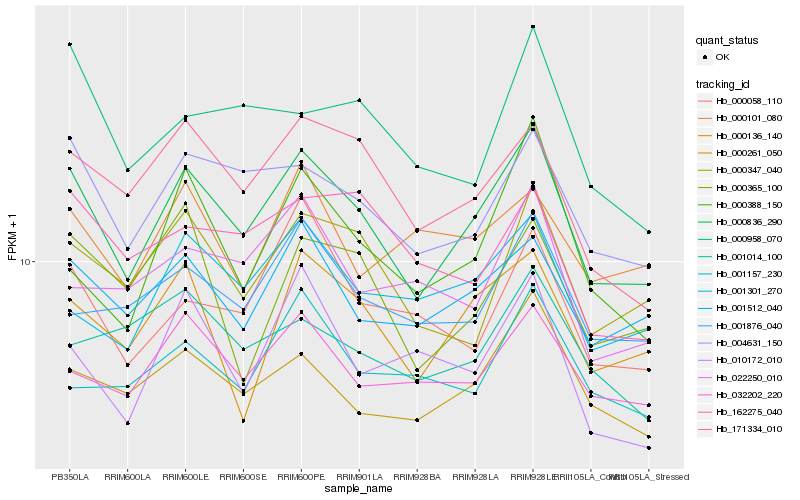
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_290 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_004631_150 |
0.0856531796 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001512_040 |
0.0913395083 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 4 |
Hb_000261_050 |
0.097069507 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 5 |
Hb_171334_010 |
0.0976684091 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 6 |
Hb_000347_040 |
0.1012643888 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 7 |
Hb_032202_220 |
0.1024674351 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000136_140 |
0.1037750694 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 9 |
Hb_001301_270 |
0.1061940495 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 10 |
Hb_001876_040 |
0.1089775638 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 11 |
Hb_000388_150 |
0.1166713373 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 12 |
Hb_000058_110 |
0.1173030844 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000365_100 |
0.1185744491 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001014_100 |
0.1230338049 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 15 |
Hb_000958_070 |
0.1230940964 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 16 |
Hb_162275_040 |
0.1231732573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001157_230 |
0.123219869 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 18 |
Hb_022250_010 |
0.1237969211 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 19 |
Hb_000101_080 |
0.1241324284 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 20 |
Hb_010172_010 |
0.1242596475 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |