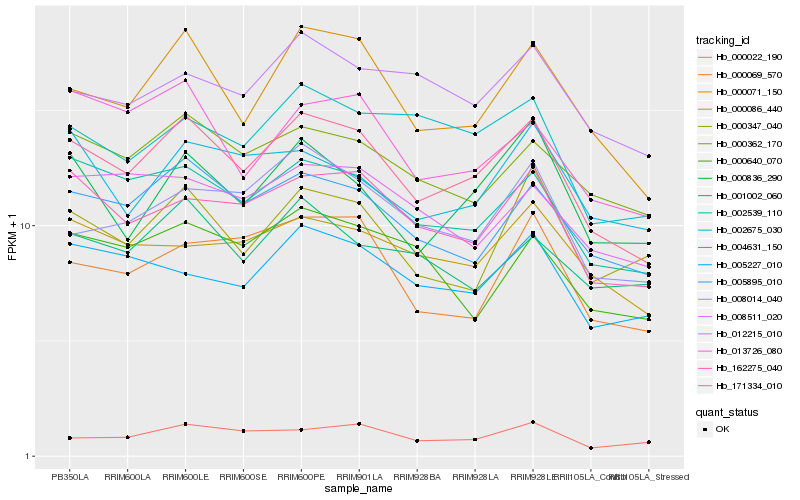
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171334_010 |
0.0 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 2 |
Hb_000071_150 |
0.0690956306 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 3 |
Hb_002539_110 |
0.0728617895 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 4 |
Hb_005895_010 |
0.07632349 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 5 |
Hb_162275_040 |
0.0795067903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000362_170 |
0.0887770097 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 7 |
Hb_013726_080 |
0.09618168 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 8 |
Hb_000836_290 |
0.0976684091 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002675_030 |
0.0981474446 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 10 |
Hb_008014_040 |
0.0982510488 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.099005478 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005227_010 |
0.099921462 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000022_190 |
0.100077517 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 14 |
Hb_000086_440 |
0.1003917238 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_008511_020 |
0.101257379 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 16 |
Hb_012215_010 |
0.1034416276 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 17 |
Hb_000347_040 |
0.1039880408 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 18 |
Hb_000640_070 |
0.105107579 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 19 |
Hb_001002_060 |
0.1061513306 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 20 |
Hb_004631_150 |
0.1068699244 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |