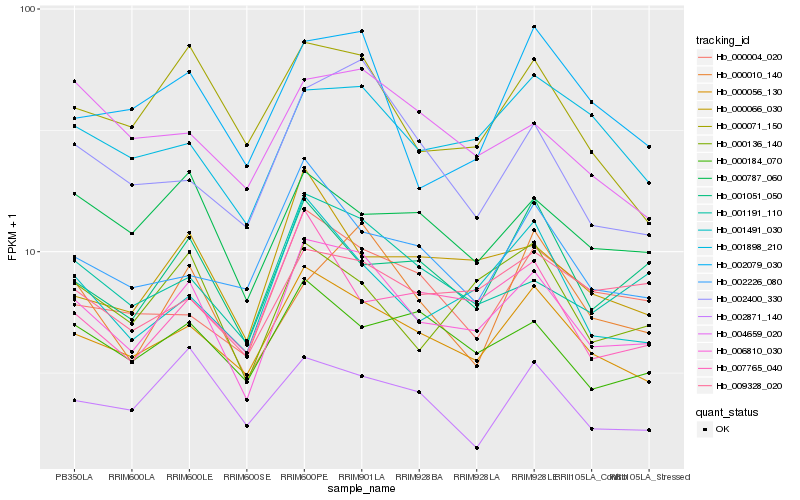
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006810_030 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_000056_130 |
0.0866044051 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 3 |
Hb_001191_110 |
0.1047322478 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 4 |
Hb_001491_030 |
0.1118524563 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000136_140 |
0.1140405843 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 6 |
Hb_000184_070 |
0.1176517666 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000071_150 |
0.1178755178 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 8 |
Hb_001898_210 |
0.1195631664 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 9 |
Hb_000787_060 |
0.1196169383 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000066_030 |
0.1225703647 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000004_020 |
0.1227205692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001051_050 |
0.1234456505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002400_330 |
0.1235141346 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_009328_020 |
0.1237748146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002079_030 |
0.128286757 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000010_140 |
0.1296642809 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_002226_080 |
0.1299087727 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007765_040 |
0.1301313394 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 19 |
Hb_004659_020 |
0.1316793165 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 20 |
Hb_002871_140 |
0.1320994462 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |