| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
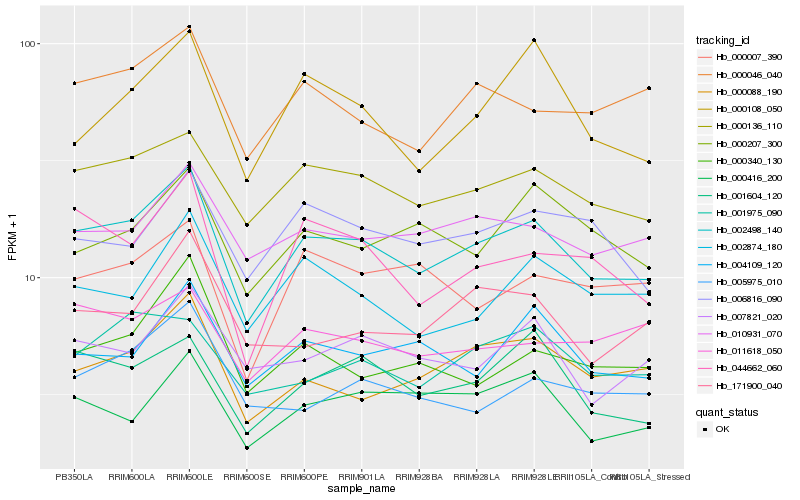
Hb_002498_140 |
0.0 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 2 |
Hb_000136_110 |
0.0715797153 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 3 |
Hb_010931_070 |
0.0920392844 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000088_190 |
0.093287232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001604_120 |
0.0950832508 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000046_040 |
0.0954904905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_044662_060 |
0.09624824 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004109_120 |
0.0984617647 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007821_020 |
0.0992867043 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002874_180 |
0.0993879039 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000416_200 |
0.1005160304 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 12 |
Hb_006816_090 |
0.1031545441 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001975_090 |
0.1032267809 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 14 |
Hb_171900_040 |
0.1038128327 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 15 |
Hb_000340_130 |
0.1045837019 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000108_050 |
0.1047133073 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 17 |
Hb_000207_300 |
0.1057957701 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_011618_050 |
0.1064837472 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 19 |
Hb_000007_390 |
0.1066624527 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 20 |
Hb_005975_010 |
0.1076654566 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |