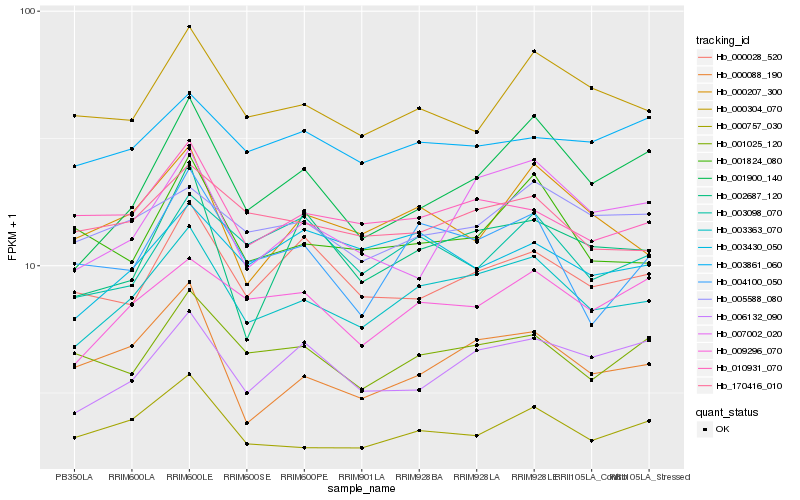
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003363_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000757_030 |
0.0789278268 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 3 |
Hb_009296_070 |
0.0831616338 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010931_070 |
0.0859599976 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_170416_010 |
0.0876415521 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 6 |
Hb_006132_090 |
0.0888783518 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000304_070 |
0.0901889044 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 8 |
Hb_005588_080 |
0.0907995545 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 9 |
Hb_001900_140 |
0.0912709512 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001025_120 |
0.0913837144 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_000088_190 |
0.0915215627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000207_300 |
0.0922425467 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004100_050 |
0.0939861615 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |
| 14 |
Hb_000028_520 |
0.0955041217 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 15 |
Hb_003430_050 |
0.0965837154 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 16 |
Hb_003098_070 |
0.0967523191 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 17 |
Hb_003861_060 |
0.0968050319 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 18 |
Hb_001824_080 |
0.0969236547 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 19 |
Hb_007002_020 |
0.0970675687 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 20 |
Hb_002687_120 |
0.0975050256 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |