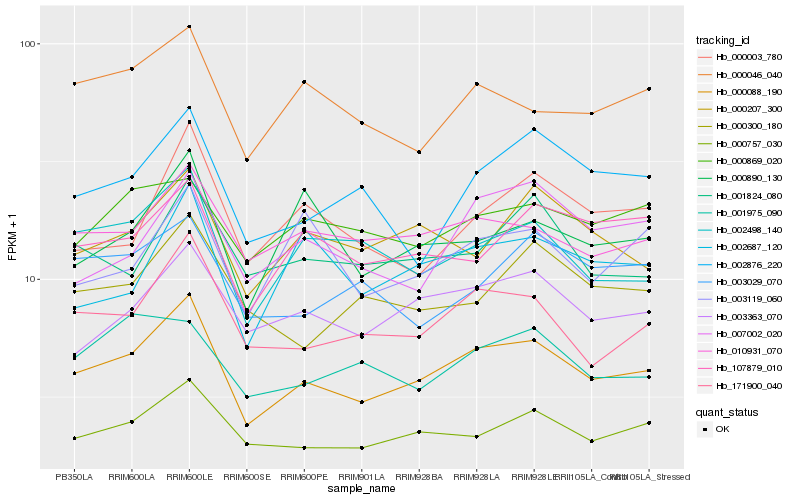
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_171900_040 |
0.0803817548 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 3 |
Hb_000757_030 |
0.0817909548 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 4 |
Hb_010931_070 |
0.0912284506 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003363_070 |
0.0915215627 |
- |
- |
- |
| 6 |
Hb_002498_140 |
0.093287232 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 7 |
Hb_000046_040 |
0.1015310665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_107879_010 |
0.1019533309 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 9 |
Hb_002876_220 |
0.1046721107 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 10 |
Hb_000207_300 |
0.1047289901 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000890_130 |
0.1052610448 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 12 |
Hb_000003_780 |
0.1066120965 |
- |
- |
hexokinase [Manihot esculenta] |
| 13 |
Hb_001824_080 |
0.1069768892 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 14 |
Hb_000300_180 |
0.1084078602 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 15 |
Hb_002687_120 |
0.1091117815 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007002_020 |
0.1091377927 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 17 |
Hb_000869_020 |
0.1093358949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 18 |
Hb_003029_070 |
0.1121726733 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_003119_060 |
0.1129714612 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001975_090 |
0.1140753674 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |