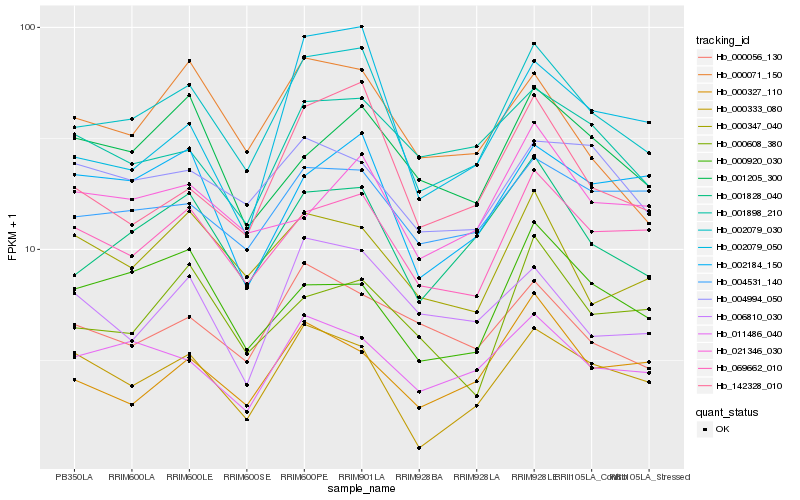
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002079_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001828_040 |
0.0823782195 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 3 |
Hb_069662_010 |
0.0935130217 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 4 |
Hb_142328_010 |
0.1030886895 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011486_040 |
0.1074931306 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 6 |
Hb_000333_080 |
0.1099726651 |
- |
- |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004531_140 |
0.1122164034 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 8 |
Hb_000071_150 |
0.1160516868 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 9 |
Hb_000347_040 |
0.1162517338 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 10 |
Hb_001898_210 |
0.1195509344 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 11 |
Hb_002184_150 |
0.1208005752 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000056_130 |
0.1213759291 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 13 |
Hb_002079_050 |
0.1228216949 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 14 |
Hb_004994_050 |
0.1233034166 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 15 |
Hb_001205_300 |
0.1264427113 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 16 |
Hb_000327_110 |
0.1277233412 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 17 |
Hb_006810_030 |
0.128286757 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_021346_030 |
0.1293191386 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 19 |
Hb_000920_030 |
0.130257948 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000608_380 |
0.1310448725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |