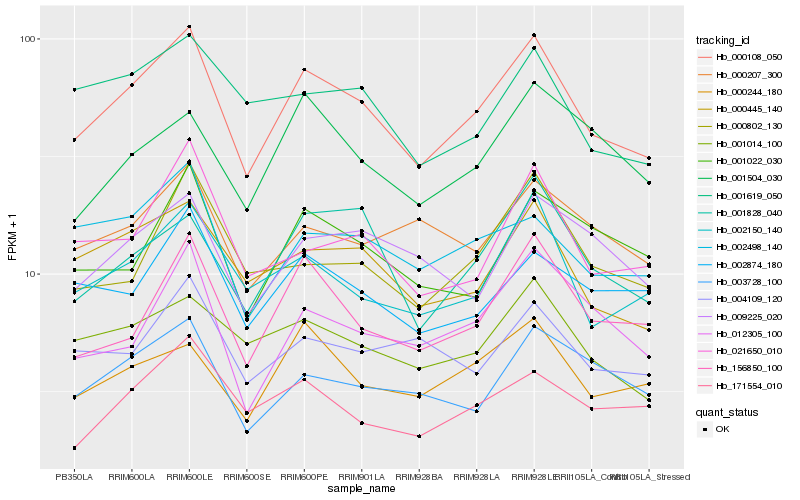
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_050 |
0.0 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 2 |
Hb_000445_140 |
0.0908135624 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 3 |
Hb_002150_140 |
0.0929741685 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 4 |
Hb_156850_100 |
0.0963001874 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 5 |
Hb_001828_040 |
0.099941675 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 6 |
Hb_001014_100 |
0.100550466 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 7 |
Hb_000244_180 |
0.101922312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003728_100 |
0.1045073463 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002498_140 |
0.1047133073 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 10 |
Hb_002874_180 |
0.105492607 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_021650_010 |
0.1065716828 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 12 |
Hb_012305_100 |
0.1073920469 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_171554_010 |
0.1085580412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 14 |
Hb_001022_030 |
0.1104390888 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 15 |
Hb_009225_020 |
0.1108284321 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 16 |
Hb_001504_030 |
0.1114571616 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 17 |
Hb_000802_130 |
0.1116721953 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 18 |
Hb_004109_120 |
0.1120023358 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001619_050 |
0.1121789383 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000207_300 |
0.1128909731 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |